Database release 2017.1

We are pleased to announce our first database release of 2017 on 26th Jan 2017. While there are no major updates in this release, it includes several bug fixes and new ligands of relevance to immunopharmacology. Our new portal, the Guide to Immunopharmacology, is now in alpha release version 3, with the first public beta release due in Spring 2017. If you would like to get involved in testing please contact us (http://www.guidetopharmacology.org/).
New contributor faculty pages
A new feature of the 2017.1 release is our contributor faculty pages. Every database contributor now has an individual page providing additional details, such as ORCIDs, home page links and subcommittee membership. We will be building on this in future releases. You can link to contributor pages by clicking on names on the main list of all contributors (http://www.guidetopharmacology.org/GRAC/ContributorListForward) or from target page contributor lists.
New ligands – 2016 drug approvals
While we generally pick up new drug approvals as they are announced, this is the time of the year when we do a cross-check against the complete list of FDA 2016 approvals (http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DrugInnovation/ucm483775.htm). This established we have 15 of the 22 entries, since we do not add anti-infectives or imaging reagents without specific target binding. As has already been alluded to in the press, this looks a really bad year compared to 2015 (https://cdsouthan.blogspot.se/2016/01/the-2015-fda-approved-small-molecule.html).
The last 2016 approval under the wire was nusinersen, an antisense ologonucleotide .
becoming the second approval of this class after eteplirsen. These breakthrough polynucleotide therapeutic modalities are of course excellent news for the benefit of patients but they do present us with particular curatorial challenges. The first of these is we cannot assign target binding data but we do briefly describe the published molecular mechanism of action in the bioactivity tab, in both these cases suppression of defective exon skipping. The next two problems are related as what formal molecular descriptors to use and how to render these as images (i.e. to produce an informative molecular picture). In a nutshell, since eteplirsen is outside the PubChem size range we have chosen Varna as an informative picture, despite the fact that two external sources (indicated in the entry) actually managed a formal rendering but produced different InChIKeys. Since nusinersen should be just inside the PubChem limit size we have both a Varna image and a SMILES string (from ChemSpider) producing a Mw of 7126 so we will check (since we will be the first submitters) how PubChem handles this.
We have already captured the first 2017 FDA approval as plecanatide for the treatment of Chronic Idiopathic Constipation (CIC) in adult patients.
It typically takes a week or so for our refreshed submissions in PubChem to go live. When the new statistics are available we will post them here.
Content fixes
Since we welcome user feedback on both navigability and content it was good to see an uptick for this in 2016. We are particularly grateful when users send us correction suggestions that we can then fix. Two cases are in this release. The first of these was a name mismatch in our olmutinib entry. The incorrect synonyms HM-71224 and LY3337641 (which refer to a blinded Hanmi BTK inhibitor) have been removed. The second was a structural error for afuresertib. We explain such fixes in the revised entries. For this and a host of other obvious reasons any and all integrators/consumers of our content are encouraged to keep on top of our new releases. We know this is a tough job, so if we can help, get in touch.
Links to SLC tables
We have added links from transporter pages to the Bioparadigms SLC Tables database. This site aggregates lots of information relevant to the Solute Carrier superfamily. We look forward to collaborating with the developers of the SLC tables in future, as their site grows. Representative example: http://www.guidetopharmacology.org/GRAC/FamilyDisplayForward?familyId=165
Database content and statistics
The number of targets stands at 2797 and ligands at 8765 with 14890 curated quantitative interactions. See http://www.guidetopharmacology.org/about.jsp.
Human targets with curated ligand interactions: 1648
Human targets with quantitative binding data: 1392
Human target with quantitative binding data to ligands with PubChem CIDs: 1265
PubMed stats
In the next release
We are working on improvements to our web services and a new version will be coming soon. The Concise Guide to Pharmacology editors and contributors are busy working on updating the concise view pages of GtoPdb with a view to releasing the updates in Spring 2017 for a new version of the Concise Guide due out in the British Journal of Pharmacology later this summer.
IUPHAR review 100 in Pharm Revs and review 21 in BJP
Two new IUPHAR reviews have been published online in January 2017.
The first is the 100th in Pharmacological Reviews, a review on the nomenclature and properties of Calcium-Activated and Sodium-Activated Potassium Channels by et al. For database entry click here.
The second is the 21st review in the British Journal of Pharmacology, an article on the evolution of RGS (Regulators of G protein signaling) proteins as drug targets by Benita Sjögren. For database entry click here.
Hot topics: The orphan GPR139 receptor is activated by peptides
GPR139 is an orphan class A G protein-coupled receptor found mainly in the central nervous system. It has its highest expression in the striatum and hypothalamus, regions regulating locomotion and metabolism, respectively, and it has therefore been suggested as a potential target for Parkinson’s disease and metabolic syndrome. Surrogate ligands have been published by Lundbeck A/S [1], Jansen R&D [2], Takeda Pharmaceuticals [3], as well as the University of Copenhagen (Gloriam group). In a new publication, the latter group describe the first combined structure-activity relationships of all surrogate agonist, and a common pharmacophore model for future ligand identification and optimization [4].
The physiological agonist of GPR139 is still elusive. GPR139 has previously been shown to be activated by the amino acids l-tryptophan and l-phenylalanine (EC50 values of 220 μM and 320 μM, respectively) [5,6], as well as di-peptides [5]. A new publication shows that the endogenous melanocortin 4 receptor agonists; adrenocorticotropic hormone and α- and β-melanocyte stimulating hormone in the low micromolar range. In addition, a potentially novel subpeptide (from consensus cleavage site) represents the most potent putative endogenous activator, so far (EC50 value of 600 nM) [7]. Together, these results indicate that GPR139 is a likely to be a peptide receptor that could act as a secondary target for melanocortin peptides or a yet undiscovered physiological ligand.
[1] Shi, F. (2011). Discovery and SAR of a series of agonists at orphan G protein-coupled receptor 139. ACS Med. Chem. Lett. 2, 303–306. doi:10.1021/ml100293q. PMID: 24900311
[2] Dvorak, C. (2015). Identification and SAR of glycine benzamides as potent agonists for the GPR139 Receptor. ACS Med. Chem. Lett. 6, 1015–1018. 10.1021/acsmedchemlett.5b00247. PMID: 26396690
[3] Hitchchock, S. (2016). 4-oxo-3,4-dihyroI-1,2,3-benzotriazine modulators of GPR139. US Patent US2016/0145218 A1. Takeda Pharmaceutical Company Limited
[4] Shehata, M.A. (2016). Novel agonist bioisosteres and common structure-activity relationships for the orphan G protein-coupled receptor GPR139. Sci. Rep. 6, 36681. doi:10.1038/srep36681. PMID: 27830715
[5] Isberg, V. et al. (2014). Computer-aided discovery of aromatic L-α-amino acids as agonists of the orphan G protein-coupled receptor GPR139. J. Chem. Inf. Model. 54, 1553–1557. doi: 10.1021/ci500197a. PMID: 24826842
[6] Liu, C. (2015). GPR139, an Orphan Receptor Highly Enriched in the Habenula and Septum, Is Activated by the Essential Amino Acids L-Tryptophan and L-Phenylalanine. Mol. Pharmacol. 88, 911–925. doi: 10.1124/mol.115.100412. PMID: 26349500
[7] Nøhr, A.C. et al. (2016). The orphan G protein-coupled receptor GPR139 is activated by the peptides: Adrenocorticotropic hormone (ACTH), α-, and β-melanocyte stimulating hormone (α-MSH, and β-MSH), and the conserved core motif HFRW. Neurochem. Int. 102, 105–113. doi: 10.1016/j.neuint.2016.11.012. PMID: 27916541
Comments by David E. Gloriam and Anne Cathrine Nøhr Jensen (Department of Drug Design and Pharmacology, University of Copenhagen)
Hot topics: X-ray crystallographic study defines binding domains for Ca2+ antagonist drugs and their molecular mechanism of action
This year witnessed a tremendous progress in our understanding of the structure-function relationship of voltage-gated Ca2+ channels. This is based on the cryo-electron microscopy structure of the rabbit Cav1.1 Ca2+ channel complex at a nominal resolution of 3.6 Å ([1] see Hot Topics Sep 20, 2016) which is now nicely complemented by a study defining the binding domains for Ca2+ antagonist drugs and their molecular mechanism of action at atomic resolution [2]. The authors took advantage of their elegant previous work solving the structure of bacterial Na+ channels (NavAb) by X-ray crystallography both in a pre-open and inactivated state [3,4]. They also engineered Ca2+ selectivity into its selectivity filter (“CavAb”, [5]) and found high affinity inhibition by the different chemical classes of Ca2+ antagonist drugs similar to L-type Ca2+ channels. Since CavAb assembles as a tetramer, this channel also replicates the four domain structure of the pore-forming subunit of voltage-gated Ca2+ and Na+-channels: an excellent model to investigate the drug-channel interaction at atomic resolution was now at hand.
As predicted for L-type Ca2+ channel α1-subunits by photoaffinity labeling studies 25 years ago [6], dihydropyridines (DHPs, e.g. amlodipine, nimodipine) bind to an extracellularly exposed intersubunit crevice formed by neighbouring S6 helices and the P-helix of the selectivity filter. Drug binding displaces an endogenous lipid molecule in this site. Interestingly, DHP binding induces a conformational changes which breaks the fourfold symmetry of the channel. As a consequence only one molecule can occupy the channel with high affinity and the Ca2+ interaction with the selectivity filter also changes. This results in a higher affinity for Ca2+ ions revealing an intriguing mechanism of action for these drugs: rather than directly blocking the pore, they enhance Ca2+ affinity for the pore such that Ca2+ itself gets stuck in the ion conducting pathway. Therefore DHPs do the opposite of what would be intuitively expected for a “Ca2+ antagonist”, namely preventing Ca2+ interaction with the channel. Instead, they allosterically enhance Ca2+ binding.
In contrast, and also in agreement with photoaffinity labeling and mutational studies phenylalkylamines (PAAs, e.g. Br-verapamil) bind in the central cavity on the intracellular side of the selectivity filter also disrupting fourfold symmetry. Since it is known that PAAs access this site preferentially when the channel opens its intracellular mouth upon activation this nicely explains their known frequency dependent inhibition. Unlike DHPs these drugs bind within the pore and thus must act as pore blockers thus satisfying the term “Ca2+ antagonist”.
This work from the Catterall lab must be regarded as a milestone in Ca2+ channel research. It not only revealed the mechanism of action of one of the most prescribed classes of cardiovascular drugs but also brings us much closer to predicting structural features of new generations of Ca2+ antagonists with high selectivity for different isoforms of voltage-gated Ca2+ channels. Within the L-type Ca2+ channel family this could be relevant for discovering Cav1.3–selective drugs as potential therapeutics for neuroprotection in Parkinson’s disease and neuropsychiatric disorders, such as autism (7).
[1] Wu et al (2016). Structure of the voltage-gated 2+ channel Cav1.1 at 3.6 Å resolution.
Nature 537:191-196. [PMID 27580036]
[2] Tang et al. (2016). Structural basis for inhibition of a voltage-gated Ca2+ channel by Ca2+ antagonist drugs. Nature 537, 117–121 [PMID 27556947]
[3] Payandeh et al. (2011). The crystal structure of a voltage-gated sodium channel. Nature 475, 353–358 [PMID 21743477]
[4] Payandeh et al. (2012). Crystal structure of a voltage-gated sodium channel in two potentially inactivated states. Nature 486, 135–139 [PMID 22678296]
[5] Tang et al. (2014). Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505, 56–61 [PMID 24270805]
[6] Catterall and Striessnig (1992). Receptor sites for Ca2+ channel antagonists. Trends Pharmacol Sci 13, 256–262 [PMID 1321525]
[7] Ortner and Striessnig (2016). L-type calcium channels as drug targets in CNS disorders. Channels 10: 7–13 [PMID 26039257]
Comments by Jörg Striessnig (Department of Pharmacology and Toxicology – Institute of Pharmacy, Universität Innsbruck)
GtoImmuPdb: technical update December 2016

Our final technical update for 2016 covers our v2.0 alpha-release, presentation at Pharmacology 2016 and future plans.
An early synopsis of the project can be found in this blog post. Previous technical blogs are available for February, MayAugust, September & November 2016.
Development Progress
Alpha-Release v2.0
Menu-bars
The menu-bars have been further development to include Processes and Cell Types. This basically extends the menu bar to have direct links to the new data types in GtoImmuPdb. The About and Resources menu items have been modified to make them specific to GtoImmuPdb. The ultimate aim of these developments is to make navigation through GtoImmuPdb user-friendly and logical. This will continue to be developed as we gather feedback.
Documentation and Tutorial
The documentation and user-guide tutorial were both updated upon v2.0 release.
Ligand List pages
We have developed the ligand list pages (which are linked to from the portal ‘ligand’ panel) to include an immuno tab that when selected lists all ligands tagged in the database as being included in GtoImmuPdb. The page now has a toggle button to switch between the GtoImmuPdb and GtoPdb views. We have also put in place a new ‘immuno ligand’ icon, to be displayed in the table with the other icons when the ligand has been tagged in GtoImmuPdb.
Ligand pages
We have extended the ligand pages to contain a new ‘Immunopharmacology’ section (with in the Summary tab). This contains any specific immunopharmacology comments specific to the ligand.
Pharmacology 2016
During December it was an privlege to be able to attend the BPS Pharmacology 2016. We not only presented a poster describing the Guide to IMMUNOPHARMACOLOGY, but were also given the opportunity to present this as a 2-miunute, one slide, flash poster presentation. The session was well attended and both the poster and presentation well received.
View presentation on slideshare
Other Development and Next Steps
The submission tool has been extended to incorporate ligand to disease associations. This is one of the first steps to fully incorporating disease association into GtoImmuPdb. These developments accompany additions to the database schema which now contains new tables to store these associations. Our expectation is to extend the schema and submission tool to also capture target-disease associations.
There are some disease terms in the database already, mostly linked to OMIM, the Disease Ontology or Orphanet. While these data resource may be adequate for annotating and describing immunological diseases and related diseases, we are exploring whether to include ICD disease classifications. Our aim is to have some GtoImmuPdb disease association in place prior to the beta-release in Spring 2017, but we are keeping this under-review.
In the next couple of months we will also be improving the current display of comments and references linked to new data types (processes and cell-types). We will also be incorporating references to the ligands tagged in GtoImmuPdb, and surfacing their display.
This project is supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
Hot topics: Will the real splice variants please stand up?
The number of alternative mRNA splice forms that map to human protein coding loci has increased to the point that nearly all proteins have such associated database records. This gives rise to the paradox that the gene build pipeline from the latest Ensembl GRCh38 reference genome assembly indicates 19,919 protein coding loci (which shrinks to 19,022 with HGNC annotation stringency) but 198,002 transcripts (i.e. nearly 10 transcripts per protein). Their is no question that a small number of these alternative splice forms, AS, (plus alternative initiations) have not only been verified to exist as proteins, have some kind of alternative biochemical functions and are also of pharmacological importance [1]. Notwithstanding, compared to the massive transcript profiling that RNAseq now provides routinely, experimentally verifying AS existence at the protein level at large scale is extremely difficult. This is because it can only be done by splice form specific antibodies, western blots detecting different size forms, top down proteomics (i.e. intact mass measurement) or the detection of alternative exon-specific trypic peptides. A recent review [2] proposes that expanding data sets from the latter approach are consistently detecting only single quantitatively dominant protein isoforms from each locus. The provocative inference is that the vast majority of the 200K odd predicted and/or verified alternative mRNA transcripts are not actually translated into proteins. This can be seen as an interesting methodological detection “gulph” between RNAseq and MS-proteomics. However, their has been previous support for the “single isoform” idea on the basis of transcript data alone [3]. An ancillary conclusion from this paper, generally overlooked in terms of its significance, was that when CDS length was taken into account approximately 50% of major transcripts did not corresponding to the ‘canonical’, max-exon, transcript as annotated in Swiss-Prot. This crucial topic is further discussed in [4].
[1] Bonner, T.I. (2014). Should pharmacologists care about alternative splicing? IUPHAR Review 4. Br J Pharmacol. Mar;171(5):1231-40. doi: 10.1111/bph.12526. PMID: 24670145.
[2] Tress et al. (2016). Alternative Splicing May Not Be the Key to Proteome Complexity. Trends Biochem Sci. Sep 16. doi: 10.1016/j.tibs.2016.08.008. PMID: 27712956.
[3] Gonzàlez-Porta et al. (2013). Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. Jul 1;14(7):R70. doi: 10.1186/gb-2013-14-7-r70. PMID: 23815980.
[4] Will the real cannoical protein please stand up.
https://cdsouthan.blogspot.se/2016/11/will-real-canonical-proteins-please.html
Comments by Chris Southan
GtoImmuPdb: technical update November 2016

During October we have made the first alpha-release (v1.0) of the Guide to IMMUNOPHARMACOLOGY. This blog post summarises some of the main features of the release and work on the documentation.
This first release marks an important step towards the public deployment of the first beta-release of GtoImmuPdb, scheduled for Spring 2017. We expect to make further alpha-releases over the next few months, as additional features are added.
An early synopsis of the project can be found in this blog post. Previous technical blogs are available for February, May, August & September 2016.
Development Progress
Alpha-Release v1.0
The portal has its own unique branding (header bar, logo and colour scheme) to distinguish it, but retains many of the layout features from the main GtoPdb site. This consistency should help users already familiar with GtoPdb to orientate themselves with the new GtoImmuPdb.

Screenshot of the GtoImmuPdb Portal, alpha-release v1.0
The portal provides a starting-point for accessing data in GtoImmuPdb, tailored to the requirements of users with a specific interest in immunopharmacology. Browsing by target, process and cell-type have been implemented in the alpha_v1.0 release. Ligands can be browsed, but there isn’t yet a immuno specific view for the results.
The portal and other pages with the GtoImmuPdb view toggled on will display a specific Guide to IMMUNOPHARMACOLOGY header and menu-bar. A consistent feature on the GtoImmuPdb pages is a ‘toggle’ button that enables the user to switch out to the standard GtoPdb view (and back).

Family page on GtoImmuPdb, showing new header and toggle button (a key feature of GtoImmuPdb)
Alpha-Release v1.0 Documentation
The main area of development over October 2016 has been to prepare the documentation for the alpha-release. These provide an explanation of the features included, how data was obtained and curated and how to use the site. Detailed release notes have been prepared, which will be incrementally added to or appended to on subsequent releases. They cover the following main sections:
- GtoImmuPdb portal
- Receptor Family pages
- Family Pages
- Detailed Target pages
- Immuno Process Association List pages
- Immuno Cell Type Association List pages
- Search
- Database Development
Documentation has also been prepared that gives details on how the data for both the process and cell type associations has been obtained. This includes a detailed spreadsheet on the full GO annotations, obtained via UniProt that form the basis of the immuno process associations.
We have also prepared a tutorial document that is a guide to navigating from the new portal, to access GtoImmuPdb data and understand the new GtoImmuPdb pages.
Alpha-Release v1.0 Data
GtoImmPdb uses the same underlying database as GtoPdb. This is has been extended to include and integrate GtoImmPdb data. The primary data-types of interest to GtoPdb, that have been addresses so far, are processes and cell-types. The database schema has been extended to accommodate these data-types and to associate them with targets in the database.
Immuno Process Data
GtoImmuPdb has defined its own set of top-level immunological process categories against which targets in the database can be annotated and which form the basis of organising, navigating and searching for immunological processes and associations.
These categories are:
- Immune system development and differentiation
- Proliferation and cell death
- Production of signals and mediators
- Regulation and responses to signals
- Migration and chemotaxis
- Cell-mediated immunity
- Inflammation
We have associated sets of Gene Ontology (GO) terms with each of these categories. This enables us to auto-curate targets annotated to any of those terms (or their children) by GO into our top-level immunological categories. GO data is obtained via an OBO file (http://purl.obolibrary.org/obo/go.obo) for the ontology, which is edited to restrict it to immuno-specific terms. We auto-curate targets to the top-level process terms by using GO annotation information from UniProt. Through UniProt, targets were selected that were annotated to the subset of GO terms and also cross-referenced in GtoPdb. This gave a total of 1,855 annotation to 401 targets.
The table below summaries the unique targets (UniProt) annotated under each category
-
GtoImmPdb ‘High-Level’ Process Distinct UniProt Immune System Development and Differentiation 124 Proliferation and Cell Death 33 Production of Signals and Mediators 74 Regulation and Responses to Signals 355 Migration and Chemotaxis 81 Cell-Mediated Immunity 99 Inflammation 261
Provision has been made in the database schema to capture curator comments against process information and annotations and the design is fully-adaptable to future changes.
Cell Type Data
The Cell Ontology provides the formalised vocabulary against which we annotated target to cell type associations. GtoImmuPdb has defined its own set of top-level immunological cell type categories against which targets in the database can be annotated and which form the basis of organising, navigating and searching for immunological cell types and associations.
These categories are:
- pro-B-lymphocytes, B lymphocytes & Plasma cells
- T lymphocytes (alpha-beta type) and their immediate progenitors
- T lymphocytes (gamma-delta type) and their immediate progenitors
- Natural Killer (NK) cells
- Polymorphonuclear leukocytes (neutrophils, eosinophils, basophils)
- Mononuclear leukocytes (syn: monocytes) (macrophages, dendritic cells, Kupffer cells)
- Mast Cells
- Innate Lymphoid Cell (added November 2016)
We have assigned one or more Cell Ontology terms to each of these categories. The assigned CO terms represents the highest level parent term(s) within the ontology for that category. For the purposes of annotation, it is these CO terms and their children that can be used when annotating a target to a given category. The exception is innate lymphoid cells which at present are not defined and included in the Cell Ontology.
Other Developments & Next Steps
Fixes have been made to out submission tool to include the ability to add/remove cell type categories and to add definitions/description of them.
Our focus in the next month is to develop the ligand browse landing pages (accessed via Ligand panel on the portal home), and add in icons to highlight immuno-flagged ligands throughout the main GtoPdb site.
We also want to develop the menu-bar navigation for GtoImmuPdb, as this will be important for the beta-release.
This project is supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
GtoPdb Ligands in PubChem
GtoPdb and its precursor IUPHAR-DB have been capturing the structures of pharmacologically relevant ligands since 2005. The fig.1. snapshot below shows the approved drug section of our eight-category ligand classification
As an active collaboration with the PubChem team, we have submitted our ligand records for every GtoPdb release since 2012. For the current release of 2016.4 the query (“IUPHAR/BPS Guide to PHARMACOLOGY”[SourceName]) retrieves 8674 Substance Identifiers (SIDs) and 6565 Compound Identifiers (CIDs). The excess of 2109 SIDs is accounted for by antibodies, small proteins and large peptides that cannot form CIDs. At just over 92 million CIDs from 473 sources, a range of property filters and full Boolean operations for combining query sets, PubChem provides an opportunity to “slice and dice” our ligand set in detailed, comparative and informative ways. A set of results is shown below.

The utilities of these intersects are outlined below (in order of counts):
- CNER refers to “Chemical Named Entity Recognition” for the automated extraction of chemistry from patents by sources submitting to PubChem (of which SureChEMBL is the largest at 16.3 million). This means that users can track-back most of our ligands to early patent filings that can often include more SAR than eventually appeared in the papers.
- Our low overlap with DrugBank indicates both sources are complementary in bioactive compound selection (i.e. the OR union is 12605)
- The possibility of sourcing purchasable compounds is important for experimental pharmacologists. From the 64 million vendor structures in PubChem we have nearly an 80% overlap and similarity searches may pick up analogues where there is no exact match.
- The “BioAssay active” tag overlaps extensively with ChEMBL entries but users can check for a range of activities for a ligand that maybe additional to the values we have extracted from selected papers.
- The MeSH term “pharmacological action” is useful but our impression is that NLM is falling behind in the PubChem indexing of this term.
- PDB ligand structures are valued database cross-references for many reasons.
- We have introduced a new feature that allows users to retrieve just our 1291 approved drug SID entries (Query “approved[Comment] AND “IUPHAR/BPS Guide to PHARMACOLOGY”[SourceName]”). The “PubChem Same Compound” select then generates 1174 small-molecule CIDs. This facilitates different types of comparative analysis between drug lists.
- As expected, our overlap with ChEMBL structures is high but we have captured 1147 structures not in this source, mainly due to different journal capture and shorter release cycles.
- The selection “unique to GtoPdb” indicates those CIDs where we are the only source in the whole of PubChem. These are predominantly novel structures we have extracted from papers but in some cases we have selected a different structure from other sources.
- There may be interest in which pharmacologically active peptides we have CIDs for. A simple Mw-cut isolates 178 entries
In regard to 7) a snapshot from our list of approved drugs is shown below

Hot topics: X-ray structure of the endothelin ETB receptor
Endothelin is a peptide that acts via two G-protein coupled receptors. ETA mainly causes vasoconstriction. In contrast ETB predominantly acts as a beneficial clearing receptor and by the release of endothelium derived relaxing factors, vasodilatation [1,2]. This paper describes for the first time the crystal structure of the endothelin ETB receptor [3]. To date less than 20 structures of Family A, GPCRs (targets of nearly half of all drugs) have been solved experimentally. The number solved for small peptides ligands are limited to the opioid receptor and the 13 amino acid neurotensin. This manuscript extends information to a much larger 21 amino acid peptide and interestingly demonstrates interaction over a substantial portion of the molecule. The authors propose a model whereby the N-terminal tail and the ECL2 β-sheet of ETB together form a lid-like architecture that covers the orthosteric pocket, predicted to form a very stable complex. This provides one structural explanation for the unusual property of ET-1 in causing long lasting responses. Mutations in ETB in receptors can result in Hirschsprung disease in humans, characterized by an absence of enteric ganglia in the distal colon and a failure of innervation in the gastrointestinal tract [2]. ETB receptor mutations are also associated with lethal white foal syndrome in horses as a result of limiting migration of melanocytes, pigment-producing cells found in hair follicles and skin.
[1] Guide to PHARMACOLOGY: ETB receptor
[2] Davenport et al. (2016). Endothelin. Pharmacol Rev. 68:357-418. PMID: 26956245
[3] Shihoya et al. (2016). Activation mechanism of endothelin ETB receptor by endothelin-1. Nature, 537, 363-368. PMID: 27595334
Comments by Anthony Davenport
Hot topics: Synthesis and SAR for depsipeptide natural products as selective G protein inhibitors
A team including the Gloriam Group at the University of Copenhagen (also the home of GPCRDB) have paper out in Nature Chemistry reporting the first total synthesis of YM-254890 and FR900359 [1] . These are related cyclic depsipeptide natural products that specifically and potently inhibit the Gq subfamily of G proteins, a relatively rare but useful and pharmacological property [3]. By a combination of solution and solid-phase approaches the team generated sufficient YM-254890 and FR900359 material for confirmation of the structures , pharmacological characterisation and the synthesis of ten new analogues of YM-254890 for SAR analysis. The paper also includes docking studies based on the X-ray crystal structure of YM-254890 in PDB 3AH8 [3]
[1] Xiong et al. (2016). Total synthesis and structure–activity relationship studies of a series of selective G protein inhibitors. Nat Chem, advance online publication, doi:10.1038/nchem.2577
[2] Schrage R, et.al. (2015) The experimental power of FR900359 to study Gq-regulated biological processes. Nat Commun. 14;6:10156. doi: 10.1038/ncomms10156, PMID 26658454
[3] Nishimura A. et. al.(2010) Structural basis for the specific inhibition of heterotrimeric Gq protein by a small molecule. Proc Natl Acad Sci; 107(31): 13666–13671. doi: 10.1073/pnas.1003553107, PMID 20639466
The two key potent ligands from the paper are included in the new GtoPdb release 2016.4. Details of this particular curation exercise are given in this blog post.
http://guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=9335

http://guidetopharmacology.org/GRAC/LigandDisplayForward?ligandId=9336
Comments by Chris Southan
Hot topics: X-ray structure of P2X3 receptor
Extracellular ATP is able to activate two families of cell-surface receptors, one of which is the ligand-gated ion channel family of P2X receptors. This family of cation channels is distinct from the remainder of the ligand-gated ion channels, as they are constructed of three (usually homomeric) subunits each with two transmembrane domains. Amongst the P2X receptors, the P2X3 is associated particularly with synaptic transmission in the sensory system and has, therefore, attracted a lot of attention as a potential target for novel analgesics and/or bladder dysfunction therapies.
In this report [1], multiple crystal structures of the P2X3 receptor are described, which allow a novel insight into the gating of a ligand-gated ion channel during the rest-agonist activated-refractory cycle, as well as with antagonist bound.
[1] Mansoor et al. (2016). X-ray structures define human P2X3 receptor gating cycle and antagonist action. Nature 538:66-71. doi: 10.1038/nature19367. [PMID 27626375].
Comments by Steve Alexander
GtoPdb database release 2016.4
We are pleased to announce our fourth database release of 2016. Version 2016.4 was published on 13th October 2016. The database is available through the Guide to Pharmacology website, download pages and web-services.
Target updates:
- GPCR updates:
- Ion channel updates:
- Other protein target updates:
Website updates
A new dendrogram visualisation of VGICs is included on the ion channel page (http://www.guidetopharmacology.org/GRAC/ReceptorFamiliesForward?type=IC). It shows a representation of the amino acid sequence relations of the minimal pore regions of the voltage-gated ion channel superfamily. the visualisation was taken from:
The VGL-Chanome: A Protein Superfamily Specialized for Electrical Signaling and Ionic Homeostasis. Frank H. Yu and William A. Catterall. Sci STKE. 2004 Oct 5;2004(253):re15. PMID: 15467096. DOI: 10.1126/stke.2532004re15
Synpharm
We have created a new sister database to the main Guide to PHARMACOLOGY – SynPharm, a database of drug-responsive protein sequences. The sequences in SynPharm are derived from interactions from the Guide to PHARMACOLOGY and using data from the Protein Data Bank. It is expected that the SynPharm database will grow as the principle Guide to PHARMACOLOGY database is updated – or indeed as further structural data is added to the PDB database pertaining to interactions already documented.
Please read the introductory SynPharm blog post (4th October 2016).
A summary of the current data can be found at synpharm.guidetopharmacology.org/about/data.
Database Statistics
In total the database now contains 14,701 curated interactions across 2,794 human targets and 8,674 ligands. More specifically, the database contain 1,429 human targets that have quantitative interactions to a ligand.
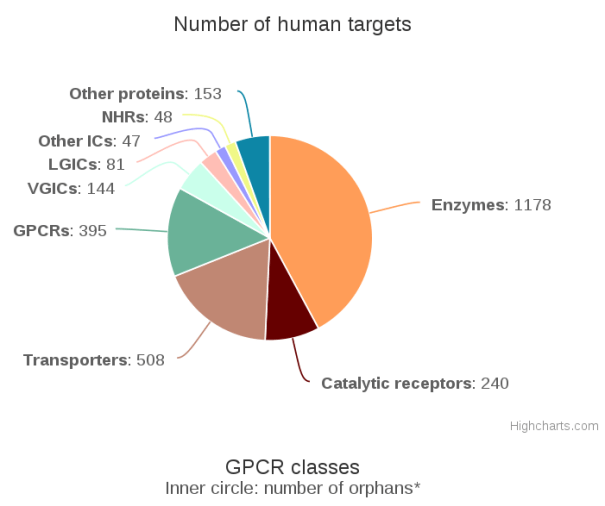
Number of human targets in GtoPdb 2016.4. Measured by number of distinct UniProt entries includes for a given target class

Breakdown of ligand classes in GtoPdb 2016.4
PubChem Links
We refresh our PubChem Substance (SID) submissions at every release and this takes a week or so to surface in their system. For 2016.4 our SIDs increased from 8612 to 8675 (if you want to execute the same query use “IUPHAR/BPS Guide to PHARMACOLOGY”[SourceName]). The same query at the Compound Identifier (CID) level increases from 6519 to 6565. As previously mentioned the 2,110 SIDs that do not merge into CIDs are antibodies, small proteins and large peptides. Note we have a slight shortfall in the CID numbers you will find listed in our ligand download lists. This is because for novel compounds where we were the first submitters to PubChem we now have to catch up with adding the new CIDs into our records.
Why Data Citation Is a Computational Problem

By Peter Buneman
The database development team encouraged me to write this off-topic blog on data citation, as it may be of interest to people involved with the IUPHAR/BPS Guide to Pharmacology (GtoPdb).
It must be almost ten years ago that Tony Harmar mentioned that he was thinking of buying digital object identifiers for the then IUPHAR database. It turned out that he was hoping that this would confer some scholarly recognition to the database, but what he really wanted to do was to get people to cite it, just as they would cite any other publication. Among other things, he wanted to ensure that the relevant contributors and curators received proper credit.
I thought about the problem for a while, wrote a rather naive paper about it, and more or less forgot about it for a few years. Then data citation became a hot topic, and with some colleagues started to think about it again. Here’s a problem: GtoPdb does a passable job of specifying the citation for each page that you see in the Web presentation, but what citation would you provide for some arbitrary SQL query on the underlying data? It turns out that this is a ubiquitous problem in data citation, and one that is tricky to solve in general.
My colleagues Susan Davidson, James Frew and I produced a general approach to this and sent it to Communications of the ACM — a publication that is widely read by computer scientists. They liked it to the extent that they made it a cover story and produced a film about it.
So thanks to Tony for the idea and thanks to the curators of GtoPdb for letting us use their database as a guinea pig.
Follow this link to the full CACM article, Why Data Curation Is A Computational Problem.
Follow this link to the video, https://vimeo.com/177314966
SynPharm: A New Annexe to the Guide to PHARMACOLOGY
We have created a new sister database to the main Guide to PHARMACOLOGY (GtoPdb) – SynPharm, a database of drug-responsive protein sequences.
Each sequence in SynPharm is derived from a GtoPdb interaction. In each case we have identified the continuous protein sequence within the receptor chain that facilitates that interaction, and provided structural, visual, spatial and affinity data.
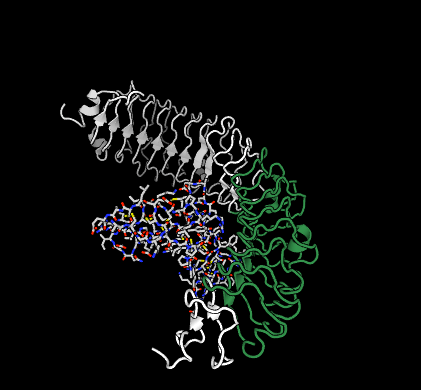
A peptide ligand (R-spondin-1) bound to its receptor (LGR4), with the bind sequence highlighted in green. See its page for more details.
Bind Sequences
Each sequence in the database represents a potentially ligand responsive protein sequence. In addition to providing a pharmacological reference as to the portion of protein chains which actually mediate their interactions with drugs, it is also hoped that SynPharm could act as a library of transferable protein modules to synthetic biologists, enabling the drug responsiveness to be conferred to a protein of choice.
In order to allow researches to assess the likelihood that a bind sequence (as the drug responsive elements are termed) will function in isolation, certain metrics are provided. We provide a ‘contact ratio’ – the ratio of internal contacts (all non-hydrogen atom pairs within the sequence within 5 Angstroms of each other, excluding atoms within two covalent bonds of each other) and external contacts (all non-hydrogen atom pairs between the sequence and the rest of the chain, less than 5 Angstroms) – and a distance matrix to show the ‘globularity’ of the sequences. Each sequence also contains a manipulable 3D visualisation of the sequence in question.

A example of a residue distance matrix. The bind sequence is represented by a dotted black line within the context of the protein chain it derives from.
In addition, we provide pages for each of the ligands that interact with a sequence, along with a small selection of the data on the ligand from the main Guide to PHARMACOLOGY database.
Creating the Data
Each interaction in the Guide to PHARMACOLOGY was mapped to one or more PDB files where possible. Some already had PDB information, and where this was not the case, the RCSB web services were queried by SMILES, InChI, name and peptide sequence (in the case of ligands) and accession number (in the case of targets) to identify more. In total, 704 interactions mapped to at least one PDB code, and after manually removing some false maps, this came down to 672. Though a relatively small proportion of the 15,000 or so interactions that GtoPdb contains, it is merely an indicator that most interactions observed have do not yet have high quality structural data.
Each interaction-PDB map was turned into a sequence by first identifying the HET code and ID of the ligand within that PDB file (generally provided by the PDB REMARK records), then identifying the residues that facilitate binding (again most PDB files already annotate this but in cases where this is not true, atomic distances were used to identify probable residues), and then using these to construct a continuous sequences. Not all maps were suitable to this – some had binding sites split across multiple protein chains, and yet more contained too many missing residues – residues flagged as missing from the crystallographic (or otherwise) experiment from which the PDB was derived. Ultimately 540 interactions had at least one PDB map that could be used to create a sequence.
It is expected that the SynPharm database will grow as the principle Guide to PHARMACOLOGY database is updated – or indeed as further structural data is added to the PDB database pertaining to interactions already documented.
A summary of the current data can be found at synpharm.guidetopharmacology.org/about/data.
GtoImmuPdb: technical update September 2016

The focus of development in the last month has been on preparing the GtoImmuPdb portal for alpha-release and building landing-pages for process and cell type association lists.
An early synopsis of the project can be found in this blog post. Previous technical blogs are available for February, May & August 2016.
Development Progress
List pages for process and cell type associations
We have developed landing pages that are reached when clicking on any of the main process or cell type categories in either of the process or cell type panel on the GtoImmuPdb portal (see Figure 1).

Figure 1: Links to process and cell type association lists pages from GtoImmuPdb portal (www.guidetopharmacology.org/immuno)
These pages list the protein targets in the database that are associated with either immunological processes or immune system cell types. In each case, the pages are split, using tabs, to show targets associated with each of the main process or cell type categories. The targets in each list are then separated by their target-class (e.g. GPCRs, ion channel, enzymes etc.). An example of the immuno process association list page is shown in figure 2.

Figure 2: GtoImmuPdb process association list page. Showing targets associated with the ‘proliferation and cell death’ category.
The table for the process associations gives the target name and family, any process association comments and lists Gene Ontology (GO) terms annotated to the target (with ID and evidence code). Two further columns show if the target has been specifically tagged as being in the Guide to IMMUNOPHARMACOLOGY by our curators, with any associated comments.
It is worth a reminder here that we auto-populate the GO annotations from UniProt. Therefore, we will see targets appearing under the process associations that have not been directly curated into GtoImmuPdb by our curators. For the time-being we will continue with the distinction between targets associated to processes (via GO only) and those that our curatorial team have identified as being of immunological relevance, and therefore directly curated as being ‘in GtoImmuPdb’.
The table for the cell type associations (see figure 3) gives the same colums as for process associations. With the exception that it lists Cell Ontology terms with their IDs.

Figure 3: GtoImmuPdb cell type association list page
Other Developments
Work has continued on implementing the site search to incorporate all new columns for process and cell type associations. This includes top-level category names, GO and Cell Ontology terms & definitions and all association comments.
A few fixes have been made to out submission tool to better handle cell type association input.
Portal development
A new logo has been added to the Guide to IMMUNOPHARMACOLOGY portal. A bespoke design by Dr. Adam Pawson. The menu bar has been adjusted to include a link to the GtoPdb home page. We have also modified some of the links within the menu-bar to keep the GtoImmuPdb focus (for target links).
Alpha-release
Our plan is to release the first alpha-version of the Guide to IMMUNOPHARMACOLOGY at the beginning of October.
This will be an internal release on our development site and will be accompanied by detailed release notes and a user guide to navigating the new pages.
This project is supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
Hot topics: High resolution structure of the voltage-gated skeletal muscle Ca2+ channel complex
In a recent article in Nature [1], Wu et al. present the cryo-electron microscopy structure of the rabbit Cav1.1 complex at a nominal resolution of 3.6 Å. Enrichment of purified channel particles without carbon film increased resolution and allowed to delineate structural features of the channel beyond those published by the authors in Science [2] six months earlier. The new structure reveals the channel in a (most likely) inactivated state (pore closed, voltage-sensors “up”), provides more complete structural detail of the α2δ-subunit and its interaction with extracellular surface of the pore-forming α1-subunit and unveils formation of a globular domain by direct interaction of the proximal C-terminal tail of α1 with its intracellular III-IV linker.
The new structural information provides new perspectives to address long-standing open questions. It will help to model human disease-related missense mutations within the Cav1.1 α1-subunit structure revealing the molecular mechanisms causing aberrant channel function, such as the formation of omega-pores in hypokalemic periodic paralysis [3]. The drug binding domains for Ca2+ channel blockers, widely used as antihypertensive drugs by blocking highly homologous Cav1.2 L-type channels in arterial resistance vessels, are highly conserved in Cav1.1. Together with recently published high resolution structure of the receptor sites for these drugs within the Ca2+-selective bacterial Na+-channel (NavAb) derivative CavAb [4], the new Cav1.1 structure will now allow to further refine the molecular details of drug interactions with L-type Ca2+ channels. The unexpected finding of a globular domain formed by the proximal C-terminus and the cytoplasmic III-IV linker of the pore subunit could provide the structural missing link for understanding how the C-terminus mediates protein kinase A regulation of the channel and controls voltage- and Ca2+-dependent channel gating in Cav1.1 and other voltage-gated Ca2+ channels.
Finally, it will be interesting to see how well the Cav1.1 α1-subunit structure was predicted by homology modeling using bacterial Na+-channels (like NavAb) or mammalian K+-channels as a template [5].
[1] Wu et al. (2016). Structure of the voltage-gated Ca2+ channel Cav1.1 at 3.6 Å resolution. Nature 537:191-196. [PMID 27580036].
[2] Wu et al. (2015). Structure of the voltage-gated calcium channel Cav1.1 complex. Science 350: aad2395. [PMID 26680202].
[3] Wu et al. (2012). A calcium channel mutant mouse model of hypokalemic periodic paralysis. J. Clin. Invest. 122: 4580–4591. [PMID 23187123].
[4] Tang et al. (2016). Structural basis for inhibition of a voltage-gated Ca(2+) channel by Ca(2+) antagonist drugs. Nature 537: 117–121. [PMID 27556947].
[5] Tuluc et al. (2016). Molecular interactions in the voltage sensor controlling gating properties of Cav calcium channels. Structure 24:261–271. [PMID 26749449].
Comments by Jörg Striessnig (Department of Pharmacology and Toxicology – Institute of Pharmacy, Universität Innsbruck)
Hot topics: Allosteric Modulation of Receptor Function and Regulation
Changeux and Christopoulos have recently described in Cell [1] how common mechanisms link the allosteric sites of activation and response within the four major receptor families of ligand- and voltage-gated ion channels, G-protein-coupled receptors, nuclear hormone receptors, and receptor tyrosine kinases. As stated in the classical “Monod-Wyman-Changeux” model [2], the signal transduction mechanism operates through the selective stabilization of the particular state to which any ligand preferentially binds. Recent research shows that these states are affected by multiple factors including oligomerization, distinct conformational ensembles, intrinsically disordered regions, and allosteric modulatory sites. These processes can be perturbed by mutations that shift the equilibrium of receptor functional states and lead to disease [3]. Conversely, marketed medicines now include a large number of allosteric modulators with the advantages of fine-tune physiological responses and offer higher on-target selectivity via more diverse binding sites [4]. Such modulators can also display increased functional selectivity through biased agonism (i.e. the association with a distinct receptor conformation and signal routing). This review summarises the unifying mechanisms for the allosteric modulation of receptor classes and provides a clear demonstration of the associated pharmacological targeting opportunities.
[1] Changeux, J.-P. and A. Christopoulos (2016). Allosteric Modulation as a Unifying Mechanism for Receptor Function and Regulation. Cell. 166(5): p. 1084-1102. [PMID: 27565340]
[2] Monod, J., J. Wyman, and J.P. Changeux (1965). On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12: p.88-118. [PMID: 14343300]
[3] Changeux, J.-P. (2013). 50 years of allosteric interactions: the twists and turns of the models. Nat. Rev. Mol. Cell Biol. 14(12): p.819-29. [PMID: 24150612]
[4] Gentry, P.R., P.M. Sexton, and A. Christopoulos (2015). Novel Allosteric Modulators of G Protein-coupled Receptors. Journal of Biological Chemistry. 290(32): p.19478-19488. [PMID: 26100627]
Comments by David E. Gloriam (Department of Drug Design and Pharmacology, University of Copenhagen).
Hot topics: Analysis of protein-coding genetic variation in humans
Lek et al. [1] in Nature, describes a tour de force large scale reference data set of high-quality protein-coding variation generated via the Exome Aggregation Consortium (ExAC) [2]. This covers 7,404,909 variants of different types that can be interrogated from an open browser set up by the team http://exac.broadinstitute.org/ [2]. Many interesting and important aspects of protein variation in both medical and evolutionary contexts are subject to statistical analysis and the results discussed. This includes loss-of function (LoF) with both clinical manifestations and consequent possible opportunities for pharmacological intervention. As just one example they investigate genetic intolerance to 179,774 high-confidence protein truncation variants (PTVd) that mapped to 3,230 highly LoF-intolerant genes. It turns out that 72% have no human disease phenotype in the OMIM or ClinVar databases. The Exac resource provides opportunities for detailed analysis of functional variation as well as a filter for analysis of candidate pathogenic variants in Mendelian diseases. The paper also indicates that most of the proposed burden of Mendelian disease alleles per-person highlighted in previous reports, is due to misclassification in the literature and/or in databases [3]. In curating target records for GtoPdb the team have been finding it increasingly challenging to select between the many sources of protein variation and different levels of supporting evidence for the phenotypic consequences thereof. On the basis of these papers and our initial assessment of their database, we would now recommend Exac as a first-stop-shop for browsing the genomic variation landscape of GtoPdb targets, with GPCRdb, Swiss-Var, ClinVar and Orphanet as orthogonal backup.
[1] Lek et al. (2016). Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291. [PMID: 27535533]
[2] Karczewski et al. (2016) The ExAC Browser: Displaying reference data information from over 60,000 exomes. bioRxiv (19 August 2016), 070581, doi:10.1101/070581
[3] Walsh et al. (2016). Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples.
Genetics in Medicine. Aug 17. doi: 10.1038/gim.2016.90. [Epub ahead of print]. [PMID: 27532257]
Comments by Chris Southan
Hot Topics: Discovery of opioid analgesics with reduced side effects
Manglik et al. [1] , writing in Nature, believe they may have found a new form of painkiller that works just as well as morphine but lacks its potentially lethal side effect. The authors have found it is not addictive by discovering a biased agonist that selectively targets the G-protein pathway over β-arrestin. Binding of agonists, such as morphine, to the μ-opioid-receptor cause very powerful reductions in the sensation of pain or analgesia via the G-protein signalling pathway but has the major side-effect of respiratory depression (the major cause of death in heroin addicts) and constipation. A further unwanted side effect limiting the use of morphine is addiction by activating the dopaminergic reward circuits. The authors show the new μ-opioid agonist PZM21 selectively activates the G-protein signalling pathway to give the desired analgesia in animal models but does not activate β-arrestin pathway, so causes little respiratory depression or constipation nor alters the dopamine pathway so would be predicted not to be addictive.
The research is important as the authors report PZM21 in mice was comparable to morphine but longer lasting. Interestingly PZM21 reduced pain in the CNS but not spinal cord in mouse models.
A biased opioid agonist TRV130 is now in Phase III trials by the company Trevena Inc that is structurally unrelated to PZM21 but has a similar pharmacological profile. Taken together, the two compounds suggest that agonists biased to the Gi/o-pathway (rather than possible differences in other pharmacological properties such as pharmacokinetics) represent a new strategy for pain control.
[1] Manglik A. et al. (2016). Structure-based discovery of opioid analgesics with reduced side effects. Nature. doi:10.1038/nature19112 advance online publication: 1-6.
Comments by Anthony Davenport
n.b. the two relevant ligands curated into GtoPdb are show below. As a new entry 9286 PZM21 will go live in release 2106.4 (September) but TRV130 was already captured as ligand 7334

GtoImmuPdb: technical update August 2016

Since our last update in May 2016 the major development extension to the Guide to Immunopharmacology (GtoImmuPdb) has been to incorporate cell type associations and develop the web-application code to display both process and cell type data.
As a reminder, a early synopsis of the project can be found in this blog post and earlier technical updates from February and May.
Development Progress
Cell Type Associations
Previously, we had written a parser to capture and populate cell type data from the Cell Ontology into the database. Since then we have determined a set of 7 high-level, immuno-relevant cell type classes (or categories), against which targets in GtoImmuPdb will be annotated. The 7 classes are as follows:
1: pro-B-lymphocytes, B lymphocytes & Plasma cells [B lymphcytes]
lymphocyte of B lineage CL:0000945
2: T lymphocytes (alpha-beta type) and their immediate progenitors [T lymphocytes (alpha-beta)]
alpha-beta T cell CL:0000789
3: T lymphocytes (gamma-delta type) and their immediate progenitors [T lymphocytes (gamma-delta)]
gamma-delta T cell CL:0000798
4: Natural Killer (NK) cells [NK cells]
natural killer cell CL:0000623
5: Polymorphonuclear leukocytes (neutrophils, eosinophils, basophils) [Polymorphonuclear leukocytes] [Granulocytes]
granulocyte CL:0000094
6: Mononuclear leukocytes (syn: monocytes) (macrophages, dendritic cells, Kupffer cells) [Mononuclear leukocytes]
monocyte CL:0000576
macrophage CL:0000235
dendritic cell CL:0000451
7: Mast cells
mast cell CL:0000097
We have assigned one or more Cell Ontology parent terms to each class. Curators will be able to annotate targets with any child terms of those parents when adding/editing cell type associations. There is also provision for free text comments about the association and the ability to include any references.
Submission Tool
The submission tool has been extended to enable the capture of cell type-target associations and related data. It has also been modified to better capture data relating to process associations (namely to include references).
GtoImmuPdb Portal

Cell type associations form in submission tool
We have continued work on the alpha-version of the GtoImmuPdb portal, and extensions to the main GtoPdb web-application to incorporate and surface GtoImmuPdb data. Previously we had implemented a toggle on target family pages to highlight targets of relevance to GtoImmuPdb. The idea behind this is so that whichever route a user takes to get to a list of targets or target families – the immuno-view can be easily switch on or off.
We have also extended the detailed target pages to display immunopharmacology comments (specific to the target), cell type associations and process associations.

Cell type and process association data being surface on the detailed target pages.
The layout of the cell type associations contains one section per each high-level cell type class. Within each section all Cell Ontology terms that have been annotated against the target are displayed, alongside comments and references.

Close-up of Immuno Cell Type Associations section of detailed target page
Similarly, the layout of the process associations has one section per high-level process class, which includes comments and references. it also includes a list of Gene Ontology (GO) Processes that are annotated to the target. These GO annotations are not input by our curators, but picked up from GO and UniProt (auto-curated). We are including the GO evidence code for these annotations.
Please note, all web-app development is only available on our restricted access test site.
Our next steps will be to improve the layout of these sections – potentially collapsing the list of GO and Cell Ontology terms (in some cases the number of terms annotated to a target can be quite high).
We will also be working on extending the code that handles our site search to include all aspects of cell type associations.
We anticipate the full alpha-release to be made in late September/early October 2016.
This project is supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
GtoPdb database release 2016.3
We are pleased to announce our third database release of 2016. Version 2016.3 was published on 21st July 2016. The database is available through the Guide to Pharmacology website, download pages and web-services. In this release, our curators have added comments to all approved drugs in the database (1,290). These comments are included in the ligand details we submit to PubChem.
Target updates:
- GPCR updates:
- Ion channel updates:
- Enzyme updates:
- Other protein target updates:
Website updates
A short, introductory video of the Concise Guide to Pharmacology has been added to the homepage. We will be bringing you more news about the concise guide through this blog in the future – so look out for those. The first, and introduction to the Concise Guide can be found here.
We have also made some minor modifications to our news, updates and announcements, consolidating these in this blog so that there is a single new feed.
Database Statistics
In total the database now contains 14,577 curated interactions across 2,789 human targets and 8,611 ligands. More specifically, the database contain 1,429 human targets that have quantitative interactions to a ligand.

Number of human targets in GtoPdb 2016.3. Measured by number of distinct UniProt entries includes for a given target class

Breakdown of ligand classes in GtoPdb 2016.3
View all the latest database content stats here.
FREE – Concise Guide to Pharmacology 2015/2016

The Concise Guide to PHARMACOLOGY 2015/2016 provides concise overviews of the key properties of over 1750 human drug targets with their pharmacology, plus links to the open access knowledgebase of drug targets and their ligands (www.guidetopharmacology.org), which provides more detailed views of target and ligand properties.
This compilation of the major pharmacological targets is divided into eight areas of focus:
- G protein-coupled receptors
- Ligand-gated ion channels
- Voltage-gated ion channelss
- Other ion channels
- Nuclear hormone receptors
- Catalytic receptors
- Enzymes and transporters.
These are presented with nomenclature guidance and summary information on the best available pharmacological tools, alongside key references and suggestions for further reading. The Concise Guide is published in landscape format in order to facilitate comparison of related targets.

Discover the FREE Concise Guide http://onlinelibrary.wiley.com/doi/10.1111/bph.v172.24/issuetoc
Watch the introductory 4 minute video on YouTube.
Hot Topics: Linking chemistry to papers
The key value of our curation is the extraction of chemistry-activity-target data from papers. Giving this relationship a formal structure in our database records not only provides direct value for users but this is also propagated globally by other databases that link to and/or subsume our content. Within the pharmacology/chemogenomic database ecosystem the largest source of chemistry <> PubMed ID links is PubChem. Many PubChem records include depositor-provided cross-references to scientific articles in PubMed, both related to chemical structures and bioassay data. The recent paper by Kim and the PubChem team [1] includes a detailed statistical analysis of these relationships that add up to 5.6 million connections between 2.2 million PMIDs and 301,000 compound records (CIDs). The paper also describes and compares in detail the different depositors, publisher-supplied and Mesh chemisty <> PMID links.
Since we are one of the PubChem depositors of these relationships, we were pleased to see not only a positive mention in this paper but also a detailed breakdown of our own contribution of 11,250 CID <> PMID relationships (presented in Table 1). Although these are small numbers compared to the total, it should be noted that ~95% of these are generated automatically (i.e. not curated) by the IBM patent extraction system that they operated on PubMed in parallel with patent document processing up to 2010. Note this chemistry-to-literature connectivity is slowly being expanded by journals, include the British Journal of Pharmacology [2].
Comments by Curation Team
GtoImmuPdb: technical update May 2016
Development of the Guide to Immunopharmacology (GtoImmuPdb) continues and this is an update of progress since our last update in February 2016. Since then, the GtoImmuPdb April Meeting was held in Edinburgh, where a detailed update on the status of GtoImmuPdb was delivered and discussions held about key points to focus over the next phase of development.
As a reminder, a early synopsis of the project can be found in this blog post.
Development Progress
Refinements have been added to the way GO biological process, of relevance to immunology, are identified and extracted from OBO files. The OBO-Edit export omitted some terms where ancestral relationship involved combinations of being ‘part-of’ something that in-turn ‘regulates’ a parent term that falls under either immune system process (GO:0002376) or inflammatory response (GO:0006954). As of 19 May 2016 the database holds 1,957 GO process terms. There are 393 targets (with cross-references to UniProt) annotated to these terms, with the total number of annotations being 1,379.
Extensions have been made to the web-application search mechanism to incorporate the high-level GtoImmuPdb process categories. These categories are: Immune system development and differentiation; Proliferation and cell death; Production of signals and mediators; Regulation and responses to signals, Cell-mediated immunity; Inflammation. The search links the GtoImmuPdb process categories to targets and is currently functional on our test site (restricted access). This needs to be extended to include GO process term, GO IDs and GtoImmuPdb process definitions.
Parsers have been developed to capture and populate cell type data from the Cell Ontology. The database now holds the cell types from the ontology, plus relationships and associations to GO processes (which will be helpful in cross-referencing). Our next steps are to determine high-level, immuno-relevant cell type classes for use on the site. A potential source are categories similar to the Immunological Genome Project; B-cells, γδT-cells, αβT-cells, T-cell activation, NK cells, myeloid cells, stromal cells, dendritic cells & stem cells. The database also needs extended further to capture target to cell-type relationships and develop associated submission tool (for editing/curation) and web-application (to surface the data to users) extensions.
GtoImmuPdb Portal
An alpha-version of the GtoImmuPdb portal has been developed (restricted access). The layout aims to compliment the GtoPdb site, whilst ensuring it is distinct through styles, logos and branding.

Early mock-up of Guide to Immunopharmacology portal.
The targets box has been developed to link to lists of targets and automatically highlight target families where there is relevance to GtoImmuPdb (this is defined by the curators by flagging targets as relevant). A toggle-button enables users to switch on/off the immuno-view. The ability to toggle the view is likely be extend to other pages. The next step will be to extend the detailed target view to display GtoImmuPdb relevant data, in the first instance general comments and process associations.

View of GPCR targets, with GtoImmuPdb toggle (blue) highlighting relevant families.
Submission Tool
Extensions have been made to enable curators to view, edit and manage associations between the high-level GtoImmuPdb Process categories and GO process terms. It also allows annotation of targets to the high-level process terms. This includes adaptations to the main webapp code that will be essential in ‘surfacing’ the process data on target detail pages. Future work will include extending to edit/manage cell type data and to enable interactions to be curated as relevant to GtoImmuPdb.
This project is supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
Recent DrugBank Changes
The DrugBank database has just announced (09 May 2016) more restrictive access conditions including user registration. Not unexpectedly, this has prompted discussion on Twitter and elsewhere (e.g. ThinkLab. including some from the DrugBank team). We enjoy long-standing contacts with the Wishart group but do not feel it is appropriate to comment per se. Nevertheless, it does seem appropriate for us to re-state our own position, and also to highlight the overlap in content between different resources. These details are not new, but have just not been juxtaposed before.
The British Pharmacological Society (BPS) has committed support for GtoPdb until 2020 and the Wellcome Trust support for GtoImmuPdb until 2018. Needless to say the management team (between, IUPHAR, BPS and the University of Edinburgh) are engaged in sustainability planning beyond those dates. We have also just applied for UK ELIXIR Node consideration.
In accordance to the commitment to openness of both funders, GtoPdb (and GtoImmuPdb when it is available) are licensed under the Open Data Commons Open Database License (ODbL) and contents under the Creative Commons Attribution-ShareAlike 3.0 Unported license. Thus, beyond appropriate attribution as a source, anyone can do anything with our content (even if we have seen minimal attributions of just a web-link to the deprecated IUPHAR-DB!). Also for the record, we have no intention of using a log-in but we do track usage and downloads since these are an important aspect of our own impact assessment.
As has been described, bioactivity databases are complex and each has unique coverage and lacunae (see http://www.ncbi.nlm.nih.gov/pubmed/24533037). We cannot therefore indicate for whom GtoPdb might at least partially substitute for DrugBank but we can point out some overlaps and differences. Firstly we should declare that, while we have been funded to capture human drug target relationships, we generally do not curate anti-infectives (although for various reasons we do have a few entries and it does not preclude doing this under new funding). Secondly, we do not annotate neutraceuticals that are metabolites. Thirdly, our drug and ligand annotation and relationship mappings have other selectivity differences to DrugBank (see http://www.ncbi.nlm.nih.gov/pubmed/26464438 and our FAQ). The upshot is that DrugBank has 7422 and we have 6293 structures with a PubChem CID entry. The overlap of 1339 thus extensively covers approved drugs for non-infectious diseases and some clinical candidates. Note also we update new drug approvals in our approximately quarterly releases (anyone needing more details on intersects and differences between the two sources is welcome to contact us).
GtoPdb database release 2016.2
The latest update of the database, version 2016.2, has been released (30th March 2016). The summary of the main updates is below. Database content in various formats can be downloaded from our download pages and accessed via web services.
This database release comes only 2 months after our last release. This has been done to coincide with the release of the IUPHAR/ASPET Pharmacology Education Site. An education portal that will be closely linked to the GtoPdb and provides high-quality training in the principles and techniques of basic and clinical pharmacology.
Target updates:
Database Statistics
In total the database now contains 14,327 curated interactions across 2,775 targets and 8,400 ligands
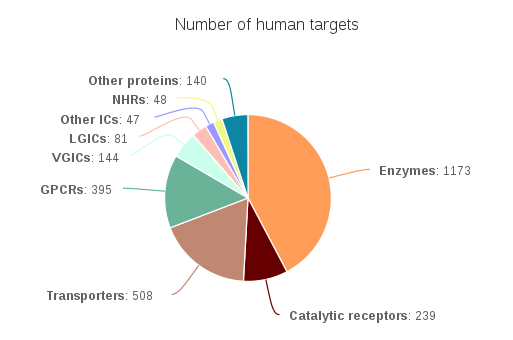
Number of human targets in GtoPdb 2016.2. Measured by number of distinct UniProt entries includes for a given target class
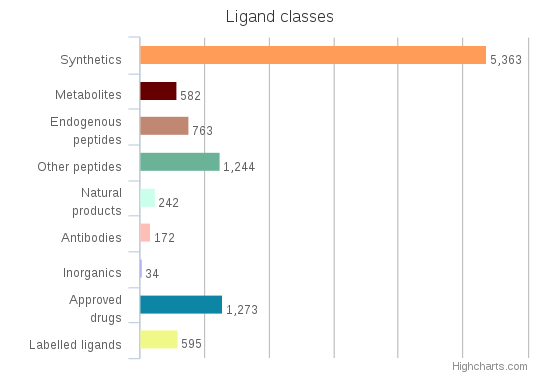
Breakdown of ligand classes in GtoPdb 2016.2
View all the latest database content stats here.
Assessment of GtoPdb in-links
A hallmark of GtoPdb is our curation of out-links as opposed to adding these by automated cross-referencing. The latter can give rise to not only false +ves and false -ves but also 1:many relationships that users find difficult to resolve. Also valuable are in-links from other relevant resources. Not only do these facilitate reciprocal navigation but also linked-data queries via inter-database web services. The Edinburgh team engages extensively with other databases at many levels, including long-standing collaborations and conference catch-ups. Indeed, a component of our value is expert selection of out-links for our GtoPdb entries, especially since pharmacology spans the domain complexity of bioinformatics, chemistry and genomics. Reciprocity of linking (a.k.a. cross-pointing) between any two databases thus becomes an enabling feature for both.
This document reviews in-links to GtoPdb from public sources that have come to our attention. However, there may be others we are not aware of (e.g. inside pharmaceutical companies). Those resources that we specifically out-link to are listed in Table 5 in our NAR article PMID 26464438. There are other cases where reciprocal in-links are under consideration but not yet instantiated (e.g. NURSA , ESTHER DrugBank and Open PHACTS).
As an open database we welcome relevant in-links. Notwithstanding, there are caveats, especially where these have been instigated without contact or our technical input. Two main problems arise. Firstly automated parsing may not be consolidated by specificity checks on their side. Secondly their update frequency my not be synchronised with our own new releases. To assess the latter we can use source entity counts and loading dates but these are not always provided. For example, we have been contacting resources we know who have not yet replaced IUPHAR-DB content by GtoPdb but its difficult to find all instances. Those in-links we know of are listed below (but please contact us if you are aware of others).
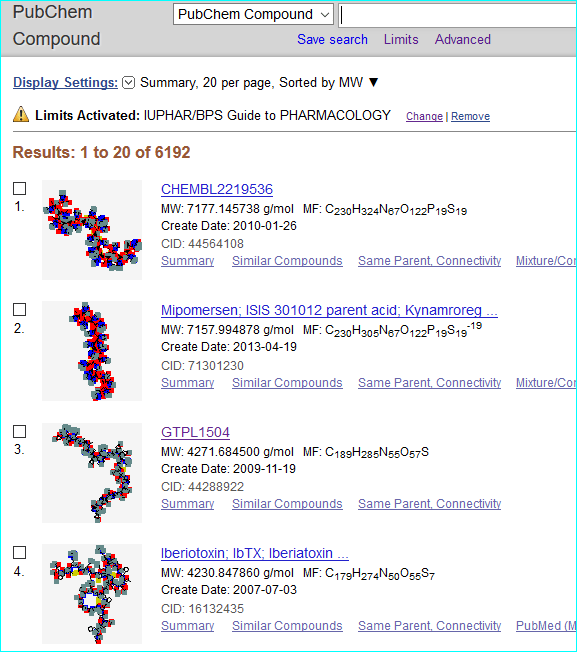
For PubChem (a global chemistry and bioactivity portal) we have 8201 ligand submissions as SIDs that each include a GtoPdb url. Of these 6192 are merged into Compound identifiers (CIDs) with a defined chemical structure. Most of the SIDs 2009 SIDs without CIDs are large peptides, small proteins and antibodies that cannot form a CID. Note we have some SID duplicates structures where we have separate GtoPdb ligand IDs pointing to radiolabel citation data without specified substitution positions that have a CID. We also have 55 BioAssay entries for 5HT sub-family chemistry mappings.
HGNC is responsible for approving unique symbols and names for human loci, including protein coding genes, ncRNA genes and pseudogenes. We have a long-standing collaboration via NC-IUPHAR. An example outlink is shown below.
UniProtKB/Swiss-Prot: We are included in the Cross-References for protein entries. These can be selected using the menu below
The query currently produces 1829 proteins with GtoPdb links as having quantitative ligand interactions.
neXtProt (PMID 25593349) is a protein-centric knowledgebase developed at the SIB Swiss Institute of Bioinformatics focused solely on human proteins. In a sense this is “forked” from Swiss-Prot but is technically distinct. It inherits our UniProt links.
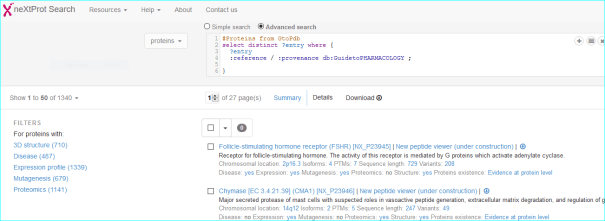
IMGT/mAb-DB is a high-quality integrated information system focused on clinical antibodies. We have a long-standing collaboration. A link example is shown below.
ChEMBL, a database of bioactive drug-like small molecules with calculated properties and abstracted bioactivities. A target link example is below.
Our ligands get a nested link in the ChEMBL interface via UniChem.
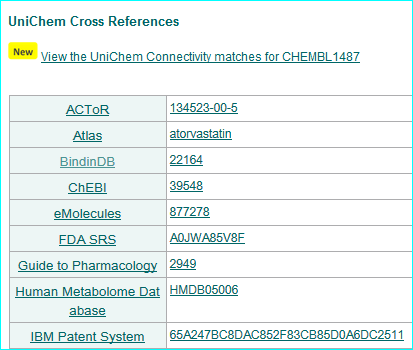
UniChem produces cross-references between chemical structure identifiers from different databases within the EBI. We are listed as a source

This was updated on 17-NOV-15 as 6006 chemical entities
***********************************************************************
MEROPS information resource for peptidases (also termed proteases, proteinases and proteolytic enzymes) and the proteins that inhibit them. An example of our cross-links is shown below.
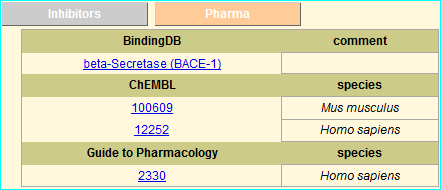
The addition of these links is specified in the latest MEROPS NAR publication (PMID 26527717)
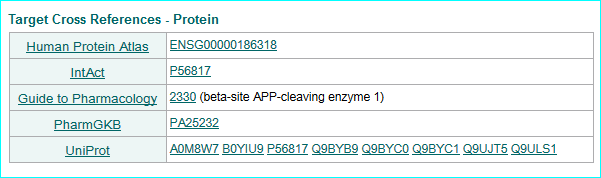
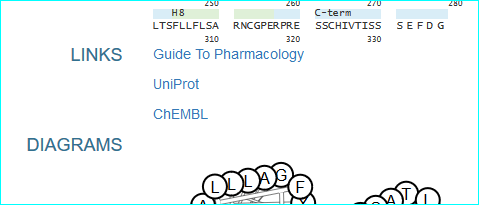
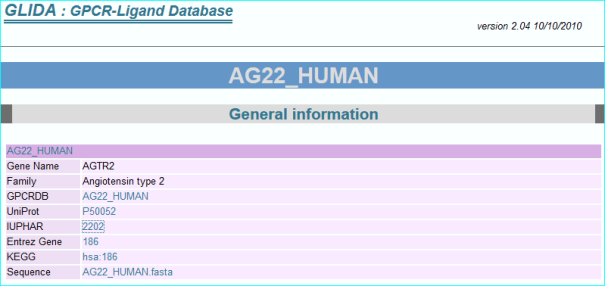
GPCRdb (in new NAR as PMID 26582914) , the information system for G protein-coupled receptors. Consequent to extensive contact and collaboration we have mentions in their paper and web resource. In addition we have pioneered reciprocal web services. A link example is shown below.
ChemSpider. This is a leading chemistry portal of 34 million compounds and also a reference structure source for OpenPhacts
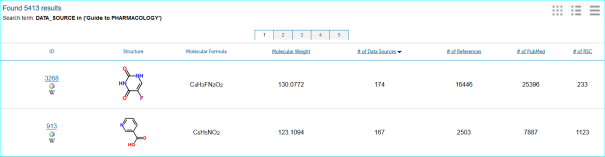
We can see from this example link for CS19973960 that we have IDs but no direct outlink at the moment.
***********************************************************************
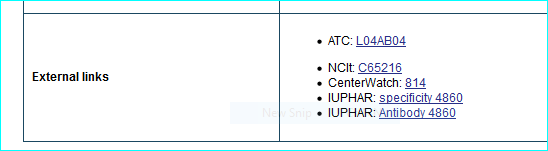
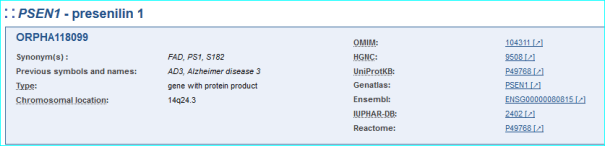
Oprhanet is the portal for rare diseases and drugs with whom we have regular contact. We are one of the eight gene-mapped IDs (see Part IV in the user guide and sample entry below)
CARLSBAD is an integrated resource based on filtered subsets from the bioactivity databases listed below (PMID 23794735).
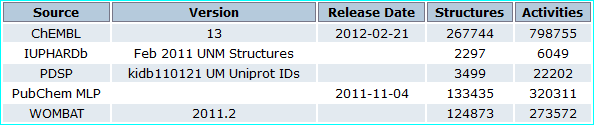
Thus the 2012 release subsumed the 2011 IUPHAR-DB set of 2297 ligands.
In the GLASS database of GPCR-ligand associations (PMID 25971743) we are cited as one their five sources. An example x-ref is shown below but the statistics are not specified.
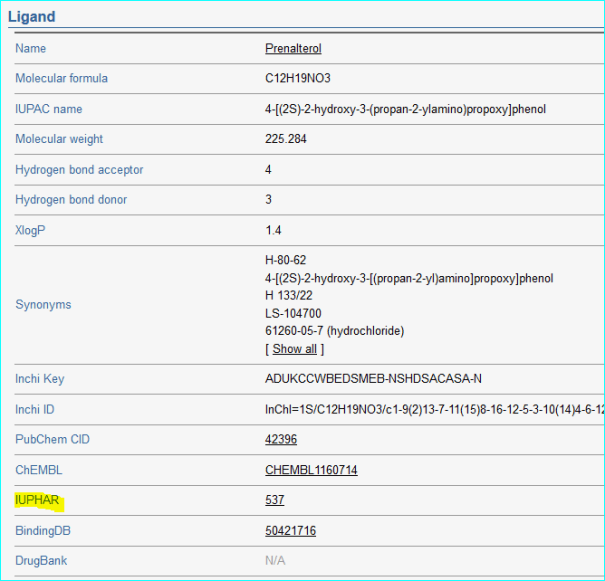
ChemProt -2.0 (PMID 23185041).
This resource with an emphasis on visual navigation in a disease chemical biology specifies eight protein < > ligand sources collated in 2012. X-ref counts were not specifies but this includes IUPHAR-DB from 2011.
The RefSeq LinkOut feature facilitates access to relevant online resources beyond the NCBI Entrez system. These include GtoPdb in the protein section with the example for NP_036236 (BACE1) shown below.

ZINC A free database of commercially-available compounds for virtual screening (PMID 26479676). The x-ref below is for IUPHAR-DB
ZINC has just undergone a major update in PubChem so our entries can be intersected
GeneCards automatically integrates gene-centric data from ~125 web sources. As we can see we became source 104 (but named IUPHAR)

We have had collaborative contact w.r.t. to their MalaCards disease database where we are source 28.

DGIdb 2.0 is a resource for mining clinically relevant drug-gene interactions (PMID 26531824). They quote on their paper “new content was imported from the IUPHAR/BPS Guide To Pharmacology (GTP) (6), accounting for 10 225 interaction claims and 1,969 druggable gene category claims” . An example of a search result specific for us, is shown below
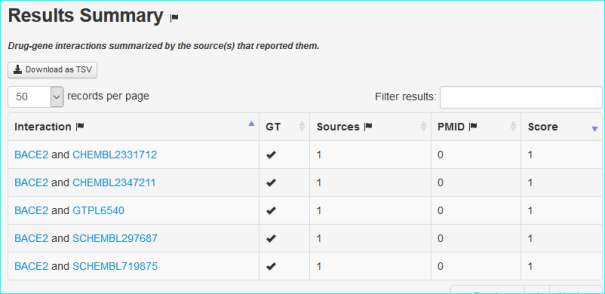
We also get a chemistry page, with the download date
And a gene page (below)
They have initiated contact and cross-mapping issues can now be addressed directly via GitHub.
We are indexed in the Protein Ontology resource that provides an ontological representation of protein-related entities by explicitly defining and showing the relationships between them.
Wikipedians have been pointing to us for some time. Examples of an established target and a more recent ligand entry are shown below.

GtoImmuPdb: technical update Feb 2016
The Guide to Immunopharmacology (GtoImmuPdb) held it’s first grant-holders meeting in London on 7th January. The meeting discussed and set-out plans for the initial phase of the project. This included discussion on new data types to include, form and content of a new homepage, which ontologies to utilise and how new data content would be generated.
An earlier synopsis of the project can be found in our previous blog post.
Some key technical decisions have been made:
- The shortened name for the database will be GtoImmuPdb.
- The GtoImmPdb will have its own homepage. This portal will provide an immunological perspective onto the database.
- GtoImmPdb will use same database as GtoPdb. It will be extended to integrate GtoImmPdb data.
- Processes/pathways – to provide a way to search data via biological processes we will be utilising target annotations to terms in the Gene Ontology (GO). There will be mapped to a simplified, GtoImmPdb specific, process list.
- Cell Types – to provide a way to search data via cell-types we will be using cell-type terminology from the Cell Ontology.
Development progress
Methods to include GO biological process terms in the database have been generated. These selected all terms under 2 parent terms, immune system process (GO:0002376) and inflammatory response (GO:0006954), resulting in a total of 1,953 terms. All these terms, plus their parent-child relationships will be included in the database and support curation and querying.
To capture an initial set of annotations of these terms against targets in GtoPdb the advanced search of UniProt was utilised. Through UniProt, targets were selected that were annotated to the subset of GO terms and also cross-referenced in GtoPdb. This gave a total of 1,364 annotation to 317 targets.
Our next steps will be to integrate this data with our front-end search mechanisms and provide access to this search via the a new GtoImmPdb portal.
Submission Tool
Minor addition to the submission tool have been made to allow curators to flag targets and ligands in GtoPdb as being relevant to GtoImmuPdb. General comments fields also add for GtoImmuPdb relevant targets.
Other planned tasks
- Domain name for portal will be guidetoimmunopharmacology.org (not yet running)
- Alpha-version of homepage to be in place by April meeting (with limited function)
- Extend GO annotation to ligands (e.g. cytokines)
Links
CiteULike Group – Immunopharmacology related to GtoImmuPdb
This project is supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
GtoPdb database release 2016.1
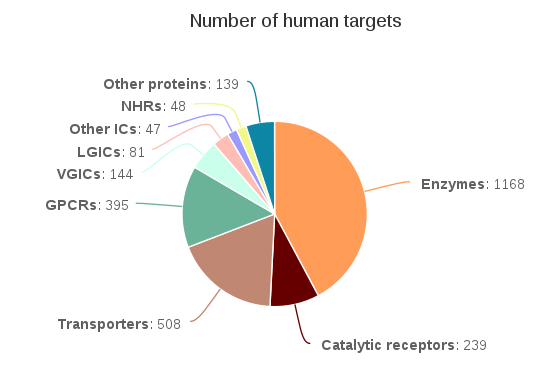
We are pleased to announce the first database release of 2016, version 2016.1 published on 4th February 2016. Below is a summary of the main updates. As always, database content, of various types and formats can be downloaded from our download pages.
Here is a summary of the latest updates:
Target updates:
- GPCR updates:
- NHR updates:
- Enzyme updates:
- Ion-Channel updates:
- Ryanodine receptor has been re-classified as a voltage-gated ion channel (previously it was a ligand-gated ion channel)
Database Statistics
In total the database now contain 14,249 curated interactions across 2,769 target and 8,328 ligands.

Breakdown of GtoPdb content by human target class

Breakdown of GtoPdb content by ligand class
View all the latest database content stats here.
GtoPdb database release 2015.3
Hot on the heels of the online advance access publication of our Nucleic Acids Research Database Issue 2016 update article we’re pleased to announce our third database release of 2015, 2015.3 published on 19th October.
Here is a summary of the latest updates:
Target updates:
- GPCR updates:
- VGIC updates:
- Members of the Transient Receptor Potential superfamily of channels
- NHR updates:
- Enzyme updates:
- Enzymes involved in hydrogen sulphide synthesis
- New ligand interactions:
- Quantitative ligand interactions added for about 150 targets which previously had no chemical modulation information in the database. As a consequence our total number of curated interactions now stands at 13859.
Website updates:
- New BLAST tool for sequence-based searching of targets available at http://www.guidetopharmacology.org/blast/. This bypasses the equivocalities of protein name searching.
View the latest database content stats here.
New project to develop “The Guide to Immunopharmacology”
We are very pleased to announce a new initiative (from 1st Nov 2015) to establish “The Guide to Immunopharmacology: Integration of targets, diseases and therapies into an expert-driven database”.
This project will be supported by a 3-year grant awarded to Professor Jamie Davies at the University of Edinburgh by the Wellcome Trust (WT).
Brief synopsis of the project
Immune/inflammatory/ infection responses and disorders have become an increasing focus of pharmacological
R&D. We will enrich GtoPdb with kinome resources linking to diseases to assist selection of new targets, tool compounds and drugs. Suggested priorities are established (JAK, PI3K, IKK) and less validated (RIPKs, IRAKs, MAP3Ks) target kinases in innate immunity.
This will later extend to adaptive immunity and kinases in selected pathogens. New data will be linked according to the existing GtoPdb expert-curation model but with a strong focus on translational aspects (e.g. clinical benefit, biomarkers and biological endpoints). In addition an immunology-orientated portal will be developed.
Co-applicants include kinase, immunity/inflammation and parasite biology experts: Dr Michael Spedding, Professor Francesca Levi-Schaffer, Professor Clare Bryant, Professor Christian Doerig, Professor Stephen Anderton, Dr Steve Alexander, Dr Doriano Fabbro and Dr Anthony Davenport. Data selection will be guided by new IUPHAR expert subcommittees set up for this task.
We owe thanks to many folk for the success of this proposal, including for their inputs to the preparation phase and letters of support (to whom we have already communicated our appreciation).
Further details will be surfaced in due course but we are also pleased that the British Pharmacological Society will continue to support the core Guide to PHARMACOLOGY resource during and after this project.
While technical decisions remain on exactly what interfaces and data structures are instantiated, we envisage both resources will be dovetailed into an expanded central database with different front-ends for users.
Any parties with Immunopharmacology interests we have not yet engaged with are welcome to make informal contact as we go forward.
Database Release August 2015
We’re pleased to announce our second public release of 2015 (version 2015.2) where we have introduced the new features and content updates listed below (and detailed on our download pages) with the objective of becoming more “consumable”. This is not only an invitation for subsumation into local systems via API calls but also via data downloads. Please note there are a number of good reasons to contact us early on in the course of any integration efforts, including technical assistance or optimisation where we can, as well as basic professional courtesy for us to know who is using our stuff. We realise some local instanciations will involve proprietory internal systems (e.g. pharmaceutical companies or competitive intelligence brokers) but we would simply like feedback on how the process (e.g. ETL procedures) went, without needing any disclosure of your internal architecture or content. We envisage 2015.3 to be ready by about November.
Target updates
- Family summary pages have been reviewed and updated in preparation for producing the Concise Guide to PHARMACOLOGY 2015/16 due out later this year
- GPCR updates:
- Dopamine receptors D2, D4 and introduction
- Vasopressin V1A and V1B
- Chemokine receptors (CCR7, CXCR1, CXCR2, CXCR3, CXCR4, CXCR5, ACKR3)
- Relaxin family peptide receptors
- Enzyme updates:
- Added inhibitors for many proteinase targets
- Mutation information, listed in the ‘Clinical mutations and Pathophysiology’ tables, has been standardised across the database. This builds upon earlier work to standardise the disease names used across the database to conform to Disease Ontology and Orphanet Rare Disease Ontology names where possible.
Content stats
| Interaction type | Count |
|---|---|
| Targets with ligand interactions | 1505 |
| Targets with quantitative ligand interactions | 1228 |
| Targets with approved drug interactions | 554 |
| Primary targets with approved drug interactions | 312 |
| Ligands with target interactions | 6796 |
| Ligands with quantitative interactions (approved drugs) | 5860 (738) |
| Ligands with clinical use summaries (approved drugs) | 1724 (1231) |
| Number of binding constants | 44691 |
| Number of binding constants curated from the literature | 13484 |
Table 1. Interaction counts. Primary target indicates the dominant MMOA.
The table shows a few of the main relationship statistics for the 2015.2 release. Further tables with target and ligand category breakdowns are available on the GtoPdb About page.
Figure 1. Relationship growth since 2012.
The first (left-most) chart shows the number of targets with curated ligand interactions while the second chart includes only those targets that are supported by quantitative data. The third and fourth charts show the number of approved drugs with data-supported targets and those that may be considered primary targets, respectively.
Website updates
We have released the first version of our REST web services for beta testing. These provide access to the data in JSON format for integration with other databases and websites. JSON is ideal for reading in with JavaScript and is easier to parse than XML. For further information and the list of web services available please see this page: http://www.guidetopharmacology.org/webServices.jsp. We welcome all feedback, bug reports, and suggestions for further development. Please try them out and let us know how you get on by emailing us. The web services offer access to lists of targets and ligands as well as specific information about targets, ligands and interactions. The web services base URL is http://www.guidetopharmacology.org/services and all target web services can be accessed with the prefix {base_url}/targets while for ligands it is {base_url}/ligands. For example, to download a list of agonists at the target GPER (GtoPdb target Id 221) with pKi >= 7 you would use this URL: http://www.guidetopharmacology.org/services/targets/221/interactions?type=Agonist&affinityType=pKi&affinity=7 For a longer list of examples see the web services documentation page.
Curating PDB links between proteins and 3D chemical structures
We know users find our curated PDB links of high value, particularly where the ligand co-crystalised within the target protein is an activity-mapped approved drug, clinical candidate or key metabolite. The comparative SAR insights these can provide are substantial, especially for those located in recent GPCR and ion channel structures. Notwithstanding, our experience indicates that verification of an explicit link (i.e. “this” chemical structure is in “this” protein sequence) is distinctly non-trivial for a range of reasons (some of which are outlined in this blog post). This means we keep our curation rules under review and carry out statistical audits of our PDB ligand content. This post is somewhat technical but what you see below is a Venn diagram that compares our ligands with two independent sources of small-molecule structure assignments (note these are derived from 3D coordinates but the analysis is actually comparing 2D structures).
The keys for the segments are as follows:
- “GtoP>PDB 293” indicates the ligands where we have curated a direct PDB link between the protein and chemical structure, counted as PubChem CIDs.
- “UC>GtoP>PDBe 938” refers to any GtoPdb ligands where (via UniChem mappings) a PDBEurope (PDBe Chem) link is recorded and have CIDs.
- “GtoP>MMDB 988” refers to any GtoPdb ligands where the PDB link is recorded via the NCBI Molecular Modelling Database (MMDB) not PDBe.
The high level interpretations are: a) We freely admit to a backlog of ~ 500 PDB ligand structures for which we could retrospectively add links. The main reason is simply a legacy effect, where older ligands have more recently appeared as PDB structures. However, some of these will be hetero-atom structures rather than specifically bound ligands or in the “wrong” targets (e.g. ACE inhibitor in Drospholia ACE protein). b) The 170 and 35 intersects are both interesting and problematic. The represent discordant PDB small molecule structures assigned either by PDBe (35) or MMDB (170) but not both (i.e. the 560).
We have three consequences going forward to enhance the database. Firstly, we have established contacts with both the UniChem and PDBe teams at the EBI (we already have one of the principal scientists from the MMDB on our chemical Curation Commitee) so we can engage with them on technical aspects. Secondly, priorities permitting, we will certainly do some back-filing of the ~ 500 missing links by triage (e.g. select newest clinical candidates first). Thirdly, we are now in a better position to cross-check any new PDB ligands for the types of discontinuities outlined above. We can then add curatorial comments and cross-pointers as appropriate.
As ever, comments on this topic are welcome.
Database Update March 2015
The latest GtoPdb update was released on March 13th and this first release of 2015 includes major updates in several areas.
GPCR updates:
- Calcitonin receptors; introduction and all receptors
- Dopamine D5 receptor
- Melanocortin receptors introduction
- P2Y1 receptor and P2Y12 receptor
- Parathyroid hormone receptors introduction
- Trace amine TA1 receptor; introduction and receptor page
- Links to BitterDB (database of bitter tasting compounds and their human receptors) added for Taste 2 Receptors
- Data on experimentally generated mutations from GPCRDB. GPCRDB is working with a consortium to collate data on GPCR mutants and is inviting submissions. Thus, the mutation data displayed here will update automatically whenever GPCRDB release new data. (See example for β2-adrenoceptor)
Kinase updates
- Added affinity data (where available) for all of the kinase inhibitors in the database (including those we previously only had large-scale matrix screening data for), tagging primary targets where appropriate
- Also added information for kinase inhibitors in clinical trials
Epigenetics target updates
- Added all of the unblinded compounds from SGC’s epigenetics compound repository
- Several new epigenetics targets added into the Other Protein Targets section
Protease updates
- This target class now includes a first (for the database) with a predrug > prodrug > drug triplet. These were curated from a paper describing how MMP12 produces its own inhibitor in a two-step activation procedure. We now have ligand entries for the peptide substrate of the protease, the prodrug and the drug.
Ligand updates
- More than 420 new ligands added in this release.
- Many new kinase inhibitors
- New monoclonal antibodies and small molecules included in pharma pipelines (including any novel drug targets)
- Sourced available binding affinity data for all monoclonal antibodies in the database using a combination of BLAST sequence analysis, patent and literature searches, tagging primary targets as appropriate
- More than 20 compounds annotated with new/expanded drug approval information
Disease information updated
- Target pathophysiology sections have been updated with standardised disease nomenclature
- Disease name synonyms added
- More links added to the OMIM and Orphanet disease databases
- Added Disease Ontology nomenclature and links to the Ontobee disease ontology browser
Website updates
- New “Specialist databases” section created to highlight database links that are of particular interest on specific target and ligand pages, for example the IMGT/mAb-DB resource for antibodies
Ligand search tools
- Our chemical structure search tool now uses the Marvin JS structure editor from ChemAxon, which replaces the older Java version. The new JavaScript version has the advantage of cross-platform compatibility
- This version should also be compatible with tablets and mobile devices
- Useful features include the ability to import molecules through various file formats, structural identifiers or by compound name
- Compounds can also be exported in many different formats or as an image file
As always, updated data files are available to download with all the new data.
21 years of NC-IUPHAR reviews and general publication updates
Update, 12 March. Inspired by the propitious event described below we have taken the opportunity to update all our publication links. This covers not only IUPHAR reviews but also papers arising from the database and additional recent pubublications co-authored by Edinburgh team members.
******************************************************************************
Last week (first one in March) the Secretary General of IUPHAR, and outgoing chair of NC-IUPHAR, Michael Spedding, informed us that 21 years have elapsed since the first NC-IUPHAR publication:
Vanhoutte PM, Barnard EA, Cosmides GJ, Humphrey PP, Spedding M, Godfraind T. (1994) International Union of Pharmacology Committee on Receptor Nomenclature and Drug Classification. Pharmacol Rev. 46(2): 111-6. [PMID: 7938161]
The direct Google Scholar retrieval for IUPHAR shows impressive metrics (including at least six articles with between 1000 and 3000 citations):
http://scholar.google.se/scholar?start=60&q=International+union+of+pharmacology&hl=en&as_sdt=1,5
Notwithstanding, in honour of the occasion, we decided to explore some additional bibliometrics. In PubMed. After some exploratory query tuning we achieved reasonable specificity with “International[Title] AND Union[Title] AND Pharmacology[Title]” which returned 96 titles:
Inspection indicated a few false positives as preceding the invited review model. So, by adding the restrict for “Pharmacological Rev.” we get a cleaner list of 90 true positives:
Note that no less than four of these (as of 2nd March) are already 2015 articles (PMIDs 5713288, 5713287, 25535277, 25287517).
To retrieve the equivalent list from the British Journal of Pharmacology, we used the query “IUPHAR[Title] AND review[Title]” to retrieve 13. However (for reasons we hope to fix), PubMed left the IUPHAR tag off review number six. Consequently the query to bring all 14 invited reviews back needs to be:
http://www.ncbi.nlm.nih.gov/pubmed/?term=IUPHAR[Title]+AND+review[Title]+OR+24428732[uid]
By making a union of the two sets (International[Title] AND Union[Title] AND Pharmacology[Title] AND “Pharmacol Rev”[Journal]) OR (IUPHAR[Title] AND review[Title] OR 24428732[uid]) we can return what looks a like a clean list of all IUPHAR reviews indexed in PubMed (you are welcome to contact us if you know of any false-negatives).
You can access these 104 via the following MyNCBI public collection link: http://www.ncbi.nlm.nih.gov/sites/myncbi/christopher.southan.1/collections/47601391/public/ (note this should update automatically for new publications). If you click on the “PubMed Commons Related Comments ” link from the facets on the LH side, you will find six articles have comments from curation team members pointing to marked-up entity links, effectively from the article to the GtoPdb. There are also some “pivots” you can do from this list to other entities indexed in the Entrez system. One of these answers the question, “how many PubMed Central articles cite (any one of) these 104 PMIDs?”. The answer is a very impressive 5978 (note these are generally only ~ 30% of the Google Scholar citations for a number of reasons). A full publication list (along with database team output) is also maintained on the GtoPdb website at http://www.guidetopharmacology.org/nciupharPublications.jsp.
The GtoPdb team would thus like to echo the congratulations to all past and present IUPHAR-invited authors.
For those less familiar with our modus operandi we can point out the “virtuous circularity” between these review articles and the content of our database. As expected, authors and subcommittee members for target families overlap extensively. Thus these authors both draw on the database for extant material to prepare a review, while also feeding updates to the curation team. As mentioned above, and in a previous post we are now instigating direct links back to the database for target and ligand entities mentioned in these reviews. Note also that committee members (or potential new ones wishing to contribute) are welcome to suggest ideas for new reviews to Eliot Ohlstein for Pharm Rev or Steve Alexander for the Br J Pharmacol.
Exploiting the new pharmacology and application to drug discovery – Abstract extension to 23 Feb
We look forward to meeting with database users, collaborators and contacts at the BPS Focused meeting 20th-21st April 2015, Edinburgh. Note that abstracts and registration are still open and bursaries for presenters available.
IUPHAR/BPS Guide to Pharmacology Team members have submitted the following titles:
“Slicing and dicing curated protein targets: analysing the drugged, druggable and tractable”
“Sorting bioactive wheat from database chaff: the challenges of discerning correct drug structures”
“Navigating between publications and databases for drug discovery: IUPHAR/BPS Guide to PHARMACOLOGY initiatives”
“Enzymes as drug targets: curated pharmacological information in the IUPHAR/BPS Guide to PHARMACOLOGY”
“Expert curated information on GPCRs in the IUPHAR/BPS Guide to PHARMACOLOGY”
“The IUPHAR/BPS Guide to PHARMACOLOGY (GtoPdb): An expert-driven knowledgebase of drug targets and their ligands”
We look forward to engaging in discussions on these themes at poster sessions and throughout the meeting, including the topics of database coverage, features, and utility for new pharmacology and drug discovery.
Team members can also be contacted at the BPS stand where we will be pleased to offer demonstrations of the database.
We also have our bi-annual (closed) NC-IUPHAR meeting over the weekend before. Thus some NC-IUPHAR committee members will attend (and be presenting at) the adjoining BPS event.
The drug class of 2014
Around this time of year there is much interest and comment from the pundits concerning the previous year’s drug approvals. Team member Chris Southan has a history of picking out aspects of the PubChem entries on his own blog and has duly done so for 2014 so these are coupled posts that pick out different aspects. As you might expect, within the team we follow up on all drug approval announcements during the year and re-tweet those of particular interest. Since we have usually already captured the agent as an advanced development candidate, we then update our entry to “approved”. We may also add new links, such as Phase 3 results, major trade names, PDB structures and cross-pointers to parent/salt splits between INNs and USANs.
What you can see in the table at the bottom of this post are the 2014 approved drug identifiers, both for the 30 PubChem CIDs and 24 GtoPdb SIDs. There are number of aspects to highlight for our own entries.
1) As explained in FAQ our current capture priorities neither extend to anti-infectives nor to large protein biologicals (other than antibodies).
2) We take diagnostics and imaging probes on a case-by-case basis. In this set we had already curated florbetaben because of its beta amyloid binding data but we have not added the Lumason contrast reagent since it has no reported pharmacology.
3) From the 41 approved drugs, eight of our entries do not have a PubChem CID. This is either because they are proteins or the peptide has not been submitted as a chemical structure (but our entries specify the amino acid sequence). However, these eight do have SIDs (pointing back to us) because we submit them to PubChem.
4) For the approved antibodies note that our entries uniquely include the quantitative target binding data where we can find this in papers and occasionally patent-only results. It thus complements the sequence records and other information you can find in the IMGT entries that we link to.
5) Note we handle drug mixtures by cross-pointing but in this set they were all anti-infectives and so are not included.
6) With the various exclusions described above we have 17 small-molecule entries for the approved drugs of 2014.
7) Updates for some of these entries will appear in the next database release.
FDA list of New Molecular Entity and New Therapeutic Biological Product Approvals for 2014
| o. | Drug Trade name | INN for parent | Date | FDA approved indication | CID | GtoPdb LID | GtoPdb SID (if no CID) |
|---|---|---|---|---|---|---|---|
| 1 | Farxiga | dapaglifozin | 01/08/2014 | Type 2 diabetes | 9887712 | 4594 | |
| 2 | Hetlioz | tasimelteon | 1/31/2014 | Sleep-wake disorder in blind individuals. | 10220503 | 7393 | |
| 3 | Vimizim | elosulfase alfa | 2/14/2014 | Mucopolysaccharidosis Type IVA (Morquio A syndrome) | |||
| 4 | Northera | droxidopa | 2/18/2014 | Neurogenic orthostatic hypotension (NOH) | 92974 | 7391 | |
| 5 | Myalept | metreleptin injection | 2/24/2014 | Leptin deficiency | |||
| 6 | Neuraceq | florbetaben F 18 injection | 3/19/2014 | Positron Emissionmography (PET) imaging of the brain in Alzheimers | 11501341 | 7769 | |
| 7 | Impavido | miltefosine | 3/19/2014 | Leishmaniasis | 3599 | ||
| 8 | Otezla | apremilast | 3/21/2014 | Psoriatic arthritis (PsA) | 11561674 | 7372 | |
| 9 | Tanzeum | albiglutide | 4/15/2014 | Type 2 diabetes | 7386 | 178103958 | |
| 10 | Cyramza | ramucirumab | 4/21/2014 | Advanced stomach cancer or gastroesophageal junction adenocarcinoma | 7390 | 178103962 | |
| 11 | Sylvant | siltuximab | 4/23/2014 | Multicentric Castleman’s disease (MCD) lymphoma | 7396 | 178103968 | |
| 12 | Zykadia | ceritinib | 4/29/2014 | Late-stage (metastatic) NSCLC | 57379345 | 7397 | |
| 13 | Zontivity | vorapaxar | 05/08/2014 | Reduce the risk of heart attacks and strokes | 10077130 | 4047 | |
| 14 | Entyvio | vedolizumab | 5/20/2014 | Ulcerative colitis and Crohn‘s disease | 7437 | 178104009 | |
| 15 | Dalvance | dalbavancin | 5/23/2014 | Skin infections | 16134410 | ||
| 16 | Jublia | efinaconazole | 06/06/2014 | Onychomycosis (fungal infection) | 489181 | ||
| 17 | Sivextro | tedizolid phosphate | 6/20/2014 | Skin infections | 11476460 | ||
| 18 | Beleodaq | belinostat | 07/03/2014 | Peripheral T-cell lymphoma (PTCL) | 6918638 | 7496 | |
| 19 | Kerydin | tavaborole | 07/07/2014 | Onychomycosis of the nails | 11499245 | ||
| 20 | Zydelig | idelalisib | 7/23/2014 | Thee types of blood cancer | 11625818 | 6741 | |
| 21 | Striverdi Respimat | olodaterol | 7/31/2014 | Chronic obstructive pulmonary disease | 11504295 | 7543 | |
| 22 | Jardiance | empagliflozin | 08/01/2104 | Type 2 diabetes | 11949646 | 4754 | |
| 23 | Orbactiv | oritavancin | 08/06/2014 | Skin infections | 16136912 | ||
| 24 | Belsomra | suvorexant | 8/13/2014 | Insomnia | 24965990 | 2890 | |
| 25 | Plegridy | peginterferon beta-1a | 8/15/2014 | Relapsing multiple sclerosis | |||
| 26 | Cerdelga | eliglustat | 8/19/2014 | Type 1m Gaucher disease | 23652731 | 7536 | |
| 27 | Keytruda | pembrolizumab | 09/04/2014 | Non-responding melanoma | 7499 | 178103956 | |
| 28 | Movantik | naloxegol | 9/16/2014 | Opioid-induced constipation in chronic pain | 56959087 | 7543 | |
| 29 | Trulicity | dulaglutide | 9/18/2014 | Type 2 diabetes. | 7638 | 223365973 | |
| 30 | Harvoni | ledipasvir/sofosbuvir | 10/10/2014 | Hepatitis C virus (HCV) genotype 1 infection | 72734365 | ||
| 31 | Akynzeo | netupitant and palonosetron | 10/10/2014 | Nausea and vomiting from cancer chemotherapy | 78759283 | ||
| 32 | Lumason | sulfur hexafluoride lipid microsphere | 10/10/2014 | Impoved contrast for echocardiograms | 17358 | ||
| 33 | Ofev | nintedanib | 10/15/2014 | Idiopathic pulmonary fibrosis (IPF) | 9809715 | 5936 | |
| 34 | Esbriet | pirfenidone | 10/15/2014 | Idiopathic pulmonary fibrosis (IPF) | 40632 | 7532 | |
| 35 | Blincy | blinatumomab | 12/03/2014 | Philadelphia chromosome-negative B-cell ALL | 7384 | 178103956 | |
| 36 | Xro | finafloxacin otic suspension | 12/17/2014 | Acute otitis externa, (swimmer’s ear) | 11567473 | ||
| 37 | Lynparza | olaparib | 12/19/2014 | Advanced ovarian cancer | 23725625 | 7519 | |
| 38 | Viekira Pak | ombitasvir, paritaprevir and rinavir tablets co-packaged with dasabuvir tablets) | 12/19/2014 | Chronic hepatitis C virus (HCV) genotype 1 infection, including cirrhosis | 86291595 | ||
| 39 | Zerbaxa | ceflozane/tazobactam | 12/19/2014 | Complicated intra-abdominal infections (cIAI) and urinary tract infections (cUTI) | 86291594 | ||
| 40 | Rapivab | peramivir | 12/19/2014 | Influenza | 154234 | ||
| 41 | Opdivo | nivolumab | 12/22/2014 | Non-responding melanoma | 7335 | 178103907 | |
| 41 entries | 30 CIDs | 25 LIDs | 8 SIDs |
MySQL relational database and other options for download
As mentioned in various places (including our FAQ) we welcome the downloading of our content in various slices and formats (listed here). Please contact us if you do this, not only out of professional courtesy but also so we can get feedback on any technical issues and/or suggested enhancements. In addition, this presents the opportunity to engage in occasional dialogue with data sources we do not yet have personal contact with.
The Guide to PHARMACOLOGY (GtoPdb) data are stored in a PostgreSQL relational database on a Linux server. For the past several releases we have made a SQL dump file available for download on our website. We have had a few requests to provide the data in MySQL format so we have produced a test MySQL version of the database migrated from PostgreSQL to MySQL. This version was created using MySQL Community Server version 5.6 on Windows and the migration was done with MySQL Workbench 6.2.
We haven’t tested the MySQL version so if you use it and find any problems please let us know by email. The PostgreSQL version is our working database; it is therefore potentially more stable than the MySQL version. If you have no technical requirement to choose one over the other, we’d recommend you stick to the PostgreSQL version. This also includes the customised text search indexes used on our website.
Note that these data are encoded in UTF-8; to use it properly with MySQL you will need to enable full UTF-8 4-byte support using the character set utf8mb4. Here is a useful post about how to do this. If you use another MySQL character set such as utf8 you may get errors with, for example, Greek characters and other symbols.
We realise that the table relationships are complex; we’re working to tidy them up our end and hope to release a new version with an annotated ERD in early 2015.
If you want a simpler slice of just the small-molecule structures and the URL pointers to the database entries (for example to integrate into a local chemistry database) the ligands.csv file may be the best choice. We do not hold SDF files but you can use the isomeric SMILES or the InChI to generate these. You may also choose to drop the rows without SMILES as these are mostly peptides and antibodies. As ever, please let us know how you get on with local integration.
Proteases in the latest GtoPdb release
The team have surfaced more fruits of our labour in release 2014.3 of GtoPdb (this is now our official acronym for the database). This post will highlight protease expansion during this curation cycle (see page 10 in our newsletter for an introduction to our Protease and Hydrolase initiative, along with the Subcommittee members). While this consolidation phase was mainly ligand additions, there are a few new targets (from pre-loaded sequences) that had compounds assigned against them for the first time (n.b. references are given in the entries so only a few are included below). Our next content iteration (i.e. from now until ~1Q15) will focus more on research target and probe compounds expansion. Note also we welcome feedback, enhancements and additions to our expanding coverage with quantitative ligand interactions.
Statistics
- Distinct UniProt protease (and MEROPS) Ids with ligand interactions = 64
- Ligand interactions (for these 64) of any type = 187 (i.e. an average of ~ 3 per target but this is a very skewed distribution)
- Quantitative target interactions (typically IC50 or Ki) = 149 (includes cross-reactivity)
- Quantitative primary target interactions with ligands = 88
- Quantitative primary target interactions with approved drugs = 39 (skewed upwards by drug-prodrug pairs where prodrug also measurable activity)
Alzheimer’s secretase targets updates:
We now have seven ligands aligned to PSEN1, our target mapping choice for inhibitors of the gamma secretase complex (n.b. a structure for the nicastrin component has just been published). These direct inhibitors are called GSIs to differentiate from “modulators”, termed GSMs.
This covers most of the declared clinical candidates. As we know, the GSIs have been having a hard time in development and if you click on the PubMed call-outs for any of the INNs (e.g. for avagacestat) you can see the stories unfolding. Notably, several GSIs are now being repositioned against tumour types thought to be driven by Notch processing (e.g. RO4929097). An interesting recent development can be followed in the links for AZ4800. This has been used for targeting amyloid β production in a mouse model for Alzheimer’s by combining with the BACE1 inhibitor AZ3979. It’s an obviously overdue experiment so kudos to AZ for publishing the first results (my guess is others with dual secretase programs have probably also tried) but, as we all know, mixture development is tough (but see the Neprilysin story below).
We have also expanded our ligand capture for BACE1 the beta secretase. Of the 12 clinical/lead inhibitors we have now curated, AZ-4217 takes the potency biscuit at 1.8 nM and has a PDB structure to boot. One of the indicators that BACE1 inhibitors might stand a better chance than GSIs in AD trials is that, since LY2886721 went down in Phase II due to liver toxicity (i.e. not target de-validation), Lilly are collaborating with AZ to take AZD3293 forward. This implies a combined level of “belief” from two experienced program teams. A serious impediment to curating (not just) BACE1 clinical leads is blinding (i.e. codes with no structures – see PMID 23159359). Not to be outdone, we have made a punt on AZD3293 from a crystallisation patent (usually a giveaway for leads) but we will see if they eventually publish and/or apply for an INN.
BACE2: diabetes target – or not?
Commercial activity around this BACE1 paralogue as a diabetes target was described in a recent patent review (PMID 23506624) and some of the compounds identified in that article are included in the six we have mapped. What remains unusual is that only a single inhibitor, Cpd J [PMID 21907142] was specified in the literature on the diabetes intervention paper in 2011, while patent filings, published since 2010 by Roche and others, have exemplified 100s of BACE2-specific inhibitors. However, three observations now cast doubt on this as a new target. First is the absence of consolidation publications from the original discovery team (i.e. not the usual string of follow-ups). Secondly, it looks like the initial flurry of BACE2-only patents has dried up, since they are now all “BACE1 or BACE2” and most are clearly bet-hedgeing BACE1 cross-screens. Thirdly, a July 2014 abstract from Novartis quotes “Taken together, these data suggest that, in contrast to previous publications, BACE1/2 inhibition does not regulate TMEM27 cleavage and has no impact on pancreatic beta-cell function and mass in diabetic mice”. Oh dear – so were Roche backing the wrong horse ridden by their academic collaborators? Let’s hope more data get published soon to resolve this target validation controversy, one way or t’other.
Neprylysin prodrug: mixture success with an angiotensin receptor antagonist
The (fixed mixture) combination drug LCZ696 from Novartis was reported to cut cardiovascular deaths by 20% vs. enalapril in a large Phase III trial. I have outlined the cheminformatic details in this previous post. The curatorial challenge is that we need to use cross-pointers rather than include mixture records that would cause all sorts of mapping problems. For this reason the GtoPdb entries for sacubatril prodrug the LBQ657 drug and valsartan each cross-point between the components internally but out to the LCZ696 mixture as CID 24755604.
PCSK9: hottest protease target on the block
After impressive early genomic target validation mAbs directed against this protein convertase to lower LDL-cholesterol levels are showing clinical success. This includes studies with the three we have curated evolocumab (Amgen) alirocumab (Regeneron) and bococizumab (Pfizer). However, therapeutic mAbs sometimes may not have their affinity parameters described in papers. This happened to be the case with bococizumab but, by searching the sequence obtained via the IMGT link, we hit the patent link and were then able to use the binding constants recorded in US8080243. What is unusual, compared to most of the proteases we have added to the database, is that apart from autocatalytic processing, the protease activity of PCSK9 is not necessary for LDL-R down regulation (PMID 22875854).
MMP12: new biology
One of the three ligands aligned against this protease is RXP470.1 is a potent and selective MMP-12 inhibitor (Ki 0.24 nM) with a 3D structure 4GQL (see below).
In 2011 the compound was reported to reduce atherosclerotic plaque development in apolipoprotein E-knockout mice. Then, a 2014 Nature Medicine paper (PMID 24784232) with one of our Protease Subcommittee members as senior author) used RXP470.1 to indicate that inhibiting extracellular MMP-12 could be a new avenue for antiviral treatments. As a sign of the times, note the other two ligands, AZD6605 and AZD1236, have both been included in repurposing lists (the latter having failed its COPD endpoints).
Cathepsin K: still going strong at 20
The sequence patent (WO9524182) priority date from early 1994 makes CATK just about the oldest genomics target that might even yet “cross the drug approval line”. After positive data from odanacatib trials, Merck intend to make their regulatory applications next year. Three other candidates are in the target entry and we have made a note to add ONO-5334 for the next release.
Cathepsin A: possible new target for heart failure
Unlike CATK, you can see from the MEROPS entry that CTSA does not have much of a drug target history. However, recent experiments by Sanofi and others with inhibitors in rodents showed reduction in cardiac hypertrophy and atrial fibrillation, implicating CTSA as a new target for heart failure (PMID 24530914). While this paper described PDB structures more potent inhibitors were published elsewhere by the Sanofi team. For the two included so far, we have used a useful curatorial convention of specifying a document link where the synonym originates from. These two cases thus become compound 8a [PMID 22861813] and example 166 (WO2014154727). While this makes for rather clunky synonym lists, without the explicit document binding, terms such as 8a or example 166 are largely useless for retrieval (n.b. if these structures eventually get development codes and/or INNs these will get promoted to the ligand names). The Sanofi patent has 177 analogues with ~ 150 IC50 data points (which makes one suspect the most potent values might have been left out!) but you can see the sub 100 nM one we picked out below.
Sanofi have added an interesting twist to the CTSA story by filing at least two repurposing “new use” patents. Usefully these were picked up in this blog post. The upshot is that Sanofi included data on CTSA inhibition by both boceprevir and telaprevir, with the latter showing a respectable 100 nM IC50.
ACE and ACE2: Updating old and new paralogues
Querying “ACE inhibitors” in PubMed returns 10352 records (as of Nov 2014) and the system generates the thumbnail graphic below (if there are enough hits).
As a consequence of the many approved drugs (listed under our ACE entry) we can see the global publication rate starting in 1981 since has fallen back after 2003 but is still significant. Evidence that the search for clinical improvements in ACE inhibition continues, despite past successes, is provided by a recent publication on the role of N-glycosylation and a crystal structure in complex with an N domain-specific phosphinic inhibitor, RXP407. In addition, other sub-domain specific inhibitors have also appeared recently, such as Lisinopril-tryptophan, an analogue of the approved drug lisinopril, which is highly selective for the C-domain corresponding to 631-1232 (n.b. there is a crystal structure for the ligand in 2X95 but in the Drosophila, not the human enzyme). There are possible clinical benefits in certain disease states for either N-selective or C-selective inhibitors.
In contrast to the older and much more studied member of the paralogous pair, querying “ACE2 inhibitors” in PubMed returns a mere seven records, starting in 2002. There is a lot of interesting literature comparing these enzymes but we have selected ligands that are more functional probes than lead compounds. The most potent of these MLN-4760 is a tripeptide with a Ki of 0.4 nM. Unusually for proteases this enzyme has a published activator in the form of XNT [PMID 18391097] that the authors suggest could decrease blood pressure, and reverse tissue remodeling.
Ubiquitin-specific protease1 (USP1) as a new cancer target:
One of the NCATS/NIH initiatives to develop molecular probes has come up with ML323 as a nM inhibitor of the USP1/UAF1 deubiquitinase complex. Having demonstrated activity in nonsmall cell lung cancer cells the team propose this as a molecular target for anticancer therapies. Our entry for this probe is shown below.

Contributed by Chris Southan
Retrospective entity linking from the literature: IUPHAR allostery review
Many of you will have noticed a gradual blurring of the distinction between publications and databases. One manifestation of this is “live linking” where entities within the text of an open access manuscript connect directly to the URL of an extrinsic database record, typically a bioactive chemical structure or a protein entry. This facilitates seamless paper < > database navigation and is complemented by paper < > paper connectivity via live-linked references. Our own engagement arose from our collaboration with the BJP and Wiley for exactly this type of markup. The results can be seen in the CGTP 2013/14 series and most recently for the NC-IUPHAR review on epigenetic pathways where a table format is used to display target and ligand links from the publication to their corresponding entries in our database.
Given the success of this, the curation team considered the possibility of being able to do this retrospectively i.e. for any relevant article that included entities potentially linkable to GtoPdb but that had no mark-up at publication time. This post describes what we came up with.
The step of listing our URLs on our blog was obvious, since we do this anyway. Less obvious perhaps, was to use the new PubMed Commons to “cross-point” interested readers between an article and our local mark-up. However, since we were already using this to spread the word about our BJP marked-up articles (since the availability of the linking is not flagged in PubMed or Europe PubMed Central), the logical connection was there.
Since it had a particularly high density of relevant entities, we chose a recent NC-IUPHAR review on allosteric ligands for our try-out. This was published in Pharmacological Reviews, which does not currently support internal linking of entities to external resources. We hope you find our listing below a useful supplement to the open access article, particularly to explore expanded relationships (e.g. ligands to other targets, or vice versa, find simillar ligands etc). These entities already existed as expert-curated entries in our database, either with interaction data or mentions in comments sections on our target pages. Note that in the publication these ligands appear within the text or tables.
| CP55940 (2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl) cyclohexyl]-5-(2-methyloctan-2-yl)phenol ) |
McN-A-343 (4-[[[(3-chlorophenyl)amino]carbonyl]oxy]-N,N,N-trimethyl-2-butyn-1-aminium chloride ) |
THRX160209 (4-{N-[7-(3-(S)-(1-carbamoyl-1,1-diphenylmethyl)pyrrolidin-1-yl)hept-1-yl]-N-(n-propyl)amino}-1-(2,6-dimethoxybenzyl)piperidine ) |
| LY2033298 (3-amino-5-chloro-N-cyclopropyl-6-methoxy-4-methyl-thieno[2,3-b]pyridine-2-carboxamide ) |
CP376395 (N-(1-ethylpropyl)-3,6-dimethyl-2-(2,4,6-trimethylphenoxy)-4-pyridinamine hydrochloride ) |
PNU-120596 (N-(5-chloro-2,4-dimethoxyphenyl)-N′-(5-methyl-3-isoxazolyl)-urea ) |
| LY2087101 (2-[(4-fluorophenyl)amino]-4-methyl-5-thiazolyl]-3-thienylmethanone ) |
xanomeline | maraviroc |
| plerixafor | 17β-estradiol | tamoxifen |
| rosiglitazone | GABA | CGP7930 |
| glutamate | CPCCOEt | ADX-47273 |
| MPEP | morphine | ticagrelor |
| cocaine | diltiazem | ethanol |
| charybdotoxin | tetraethylammonium | ivermectin |
| verapamil | ω-agatoxin IVA | ω-conotoxin GVIA |
| astemizole | dofetilide | tetrodotoxin |
| batrachotoxin | saxitoxin | 3,3,5-triiodothyroacetic acid |
| tretinoin | GW0072 | oxotremorine-M |
| alcuronium | cinacalcet | diazepam |
| strychnine | gallamine | glycine |
| picrotoxin | aniracetam | cyclothiazide |
| flumazenil | ketamine | FGF-1 |
| IL-1β | pertuzumab | trastuzumab |
| GS39783 | etomidate | propofol |
| ifenprodil | rapamycin | FITM |
| galantamine | iperoxo | amlodipine |
| desflurane | flurazepam | gevokizumab |
| zopiclone | hanatoxin |
The following ligands from the article are new entries in the database
| CX614 (2H,3H,6aH-pyrrolidino(2,1-3′,2′)1,3-oxazino(6′,5′-5,4)benzo(e)1,4-dioxan-10-one ) |
TCN-201 (3-chloro-4-fluoro-N-[4-[[2-(phenylcarbonyl)hydrazino] carbonyl]benzyl]benzenesulfonamide) |
LUF6200 |
| SSR1281129E (sodium 2-amino-5-(1-methoxy-2-methylindolizine-3-carbonyl)benzoate ) |
Org27569 (5-chloro-3-ethyl-N-(4-(piperidin-1-yl)phenethyl)-1H-indole-2-carboxamide) | CGP13501 |
| bisphenol A | andarine |
A number of targets were also detailed in the paper, at which the ligands act allosterically. The table of links below directs readers to the corresponding entries on our database. The nomenclature used in the table is the same as in the publication.
Contributed by Chris Southan and Helen Benson
P.S. It was interesting to see that PubMed Commons picked us up for a highlight
A Pharmacologists’ Guide to Resolving Chemical Structures and their Protein Targets from the Literature
This is an introduction to resolving bioactive ligands and their protein targets from the literature. It includes their conversion to standardised molecular identifiers so these can be communicated to the Guide to PHARMACOLOGY (GtoPdb) team.
(n.b. identifying what is already captured in the database is addressed in this tutorial).
The utility is conceived for three groups:
1) NC-IUPHAR Committee members updating target families in GtoPdb;
2) Authors and publishers who are (or considering) incorporating GtoPdb links into manuscripts;
3) GtoPdb users who would like to directly recommend new content (including their own papers).
This outline extends from the minimal requirement of identifying the document, to a complete set of specifications that can be directly entered into the database. It is envisaged that contributors can send us comments and cross-references that extend the basic identification. The curation team will then select entries for curation but we use the comment fields to add contextual annotation. This expert distillation adds unique value to the database.
While we do not claim perfection, GtoPdb exemplifies not only comprehensive entity resolution but also highlights ambiguity (e.g. in curator notes). This introduction assumes basic familiarity with chemical structures and protein sequences. In addition, we recommend pharmacologists increase their familiarity with PubChem and UniProt in general, since these are key resources that GtoPdb links out to (n.b. molecular resolution is important both to extend the understanding of the papers you read and those you might write or referee).
The concept of reading a journal paper “D” that describes an assay “A” with a quantitative result “R” for compound “C” that modulates the activity of a target protein “P” is familiar. While it does not encompass all permutations, the relationship can be expressed in shorthand as “D-A-R-C-P” (PMID 21569515). The GtoPdb curation team have converted 1000s of these relationship chains into structured database records. The shorthand can also describe variants, such as A-R-C-P for PubChem Bioassay or matrix screens (i.e. without D since A-R is web-based), C-P for DrugBank-type target annotation (i.e. no direct D-A-R) and agents with unknown mechanism as D-A-R-C (i.e. no P). Inspection of different GtoPdb entries gives an idea of how we populate database records and practical ways of resolving the ligand and target (i.e. C and P) entities are described below. Importantly, identifications are best communicated to us as URLs since this is less error-prone (i.e. rather than typing out a chemical or protein ID number, just copy-and-paste the primary URL). As an example, we can use a paper on the AstraZeneca clinical development compound AZD9668.
Document. While we can resolve the A-R-C-Ps within a new journal paper, the more our collaborators can do in advance the better. Thus, a PubMed number (PMID) for “D” is convenient since this can be a URL. A DOI will also do (but check it works please). While a traditional citation is acceptable, this should include the full title. Googling this is likely to find a URL from PubMed, PubMed Central, EuropePMC or the publisher’s site with a DOI. The curation team also have CiteUlike accounts so we would be pleased to accept input in this way, since copying over a reference is thereby error-free, automatically out-linked and gives you the facility of adding collaborative notes (including entity identifiers). Note also we include links to some non-journal “D” types, but we expect you to judge the provenance of non-peer reviewed sources. Patent numbers are useful but you will need to establish that the document actually contains A-R-C-P relationships (but we can advise on this). As a search term in PubMed, AZD9688 returns 9 papers but the first of these “AZD9668: pharmacological characterization of a novel oral inhibitor of neutrophil elastase” is a likely candidate for extraction of the primary data for this lead compound.
This “D” could thus be communicated to us as either;
- PubMed URL
- Publisher’s version
- DOI 10.1124/jpet.111.182139
- Your own CiteUlike library reference (preferably with notes added) or copy over to our GtoPdb Triage group library
Assay. The specifications of assay conditions are not extensive in GToPdb, since we provide the reference wherein these are described. However, it is useful to us if you highlight relevant sections from the paper (or paste into CiteUlike notes). This helps us specify the molecular mechanism of action (mmoa), either implicitly (e.g. the target is identified in a different section of the document) or explicitly (e.g. competitive inhibition or partial agonism is specified). In the AZD9688 example conditions are described in Materials and Methods, The section starts with “The potency and selectivity of AZD9668 were determined by measuring the cleavage of peptide substrates to products by a range of serine proteases…” Note that extracted summary assay descriptions and results from the same paper may surface in multiple places on the web (e.g. ChEMBL, PubChem BioAssay BindingDB and BRENDA). These cannot be expanded on here but they can differ in descriptive text.
Result. This needs to be expressed as a standard parameter such as IC50 EC50, Ki, Kd etc. We typically report these as written in the paper, but convert concentrations to nM and then log these to pAct (so if you come across ug or ng please do the nM conversion). Note we round down reported results where they clearly exceed the significant figures appropriate to the experiment (i.e. typically two or three). In the AZD9688 paper we find a detailed description“AZD9668 had a high binding affinity for human NE (KD = 9.5 nM) and potently inhibited NE activity (Table 1; Fig. 2). The calculated pIC50 (IC50) and Ki values for AZD9668 for human NE were 7.9 (12 nM) and 9.4 nM, respectively. In addition Table 2 includes a log transformed Standard Error of the Mean as a pIC50 of 7.9 ± 0.12.
Compound. Public database chemical identifiers (e.g. PubChem, ChemSpider, ChEBI, DrugBank or ChEMBL) or commercial ones (e.g. CAS registry numbers) are not yet widely used in papers. For medicinal chemistry journals in particular, structures can be obscurely labelled in SAR tables as “11h”, or even as Markush-type enumerations. However they are usually made explicit via structure images in Results and/or as IUPAC names in M&M. Where names are used for “C” the following are useful a) a shorthand biochemical name b) a company code name c) an INN or d) an English/US brand name. For a PMID MeSH can be a useful first-pass for entity resolution. You can inspect this by opening up the terms under the PubMed text and also see what is linked under “Related information” on the lower right of the page. The links for the AZD9688 paper are shown below.
Exploiting MeSH within the NCBI Entrez system cannot be expanded here. However, familiarisation helps you with entity nomenclature choices (since it is part of what MeSH annotators do) but the specificity is somewhat patchy due to various types of false-positives and false-negatives. Notwithstanding, this paper is a successful example, since the PubChem compound link takes you directly to CID 46861623
The resolving operations for chemistry can be termed name-to-structure (NTS). This is particularly challenging where queries in chemistry sources retrieve different structures for the same text name, or in the case of company code names, sometimes no structure at all (see PMID 23159359). However, in PubChem, an empirical guideline is if a large number of submitters agree on the structure (i.e. many substances merged in the CID) and the names in multiple sources match, both strucuture and name may be correct. If you draw a blank in PubChem you can try the NCBI “all database” Entrez search (e.g. with a code name) that includes PubMed and PubMed Central. Note that Google Scholar may give true hits not found in NCBI Entrez since it indexes text from behind some publishers paywalls. Those of you with access to SciFinder may occasionally find NTS matches (performed as a concept search) not in Entrez or Google Scholar.
Good papers elinate the equivocality of “C” by descriptions of either a) a 2D image, b) IUPAC systematic name (sometimes in supplementary data) c) SMILES, d) InChI string or e) an InChIKey. You can send us any of these but it is better to resolve them to database IDs yourself by any of the following routes;
- Search PubChem via SMILES, InChI string, InChIKey, .SD or .mol file
- SciFinder search with SMILES or InChI string to resolve a CAS No.
- InChIKeys can be rapidly checked in Google (PMID 23399051)
- IUPAC names converted to structures using chemicalize.org or OPSIN
- Images extracted to structures with OSRA.
- Use a chemical sketcher that outputs a structure
The AZD9668 paper is an easy case, since it exemplifies the structure as an image, an IUPAC name, along with the INN link for alvelestat.
As an example, we can corroborate the structure by automated conversion of the IUPAC name using chemicalize.org.
These two routes to chemical entity NTS resolution concur with a third result, a direct PubChem search for “AZD9668” shown below.
Protein. The use of standardised names and accession numbers to enable the resolution of “P” is more common than for “C” entities in papers. Nevertheless, ambiguity can still be a problem. As you can see in GtoPdb, we cross reference commonly used protein or gene identifiers, including our own NC-IUPHAR nomenclature. For many reasons we use the UniProt ID as an unequivocal, species specific, primary identifier, which, for human proteins, is the Swiss-Prot ID. We can also use the approved HGNC symbol. For those more familiar with the NCBI-world of annotation, a RefSeq NM or NP is aOK for us, but an Entrez GeneID is preferable. In cases where the paper specifies a protein complex you may be able to resolve this to the constituent subunit IDs (but we can check this). In the AZD9688 paper the protein name “Neutrophil Elastase” can be resolved to UniProt P08246.
It can also be identified as HGNC symbol ELANE, the Entre Gene entry 1991 and our own GtoPdb protein ID 2358. Note, as for many enzyme families with a history of alternative names, ELANE can be confused with five members of the chymotrypsin-like elastase family (CELA1, CELA2A, CELA2B, CELA3A and CELA3B). This includes what used to be called pancreatic elastase, that now splits into the last three gene names in the list.
Splice-variant specific pharmacology presents another challenge for molecular resolution (see PMID 24670145). For papers where this is highlighted, an iteration with the curation team is advised but note that Swiss-Prot includes published alternative splicing as numbered isoform cross-references with specific URLs. For a publication, the key question for annotating differential pharmacology (e.g. significantly different IC50 for splice forms) is that the experimental characterisation needs to be assignable to a single sequence-defined splice form.
Appendix I Direct entity mark-up from publishers
As exemplified in the British Journal of Pharmacology publications from NC-IUPHAR (e.g. PMID 24528243) the PDF and HTML versions include live links to GtoPdb for ligands and targets. Analogously, some journals provide direct links to PubChem. The first of these was Nature Chemical Biology that has now been followed by Nature Chemistry and Nature Communications. Some Elsevier journals now also include PubChem CID numbers and links.
Appendix II An outline of key structural specifications for chemical entities
 Note that most chemistry tool-kits can execute the interconversions indicated by the arrows and major chemistry databases will pre-compute links between them. However, the round-tripping may not be perfect.
Note that most chemistry tool-kits can execute the interconversions indicated by the arrows and major chemistry databases will pre-compute links between them. However, the round-tripping may not be perfect.
Appendix III Peptides and radioactive analogues
Pharmacologically active peptides (as a “C” entity) present different NTS problems to small molecules. They require options of representation as three-letter codes or FASTA character string. In addition, endogenous unmodified peptides specifically cleaved from precursors are usually specified in the Swiss-Prot features lines (so you can send us the URLs for these). Many are also specified as SMILES in PubChem CIDs. Complications arise from names that may be not be IUPAC standard and/or include posttranslational modifications such as N-acetylation or C-amidation. Exogenous synthetic peptides have the same issues and may need to be iterated with us. Radioactively labelled analogues (small molecules or peptides) also feature prominently in the pharmacological literature. However, sometimes the molecular position of the label is unspecified. Here again, we have resolving approaches where may find them in PubChem but it would be useful if you have a suppliers catalogue reference.
Appendix IV Bulk extraction
A lot of time may be saved where compounds and proteins in a publication can be found either already marked-up and/or they can be extracted in bulk (i.e. for the entire document or sections therein) but only a few options can be noted here. PubMed Central runs its own Entrez look-up tagging for full text articles. Other automated options for genes and proteins include EBI Whatzit and Utopiadocs. For small molecules chemicalize.org is the most effective, particularly for IUPAC names. Note that Europe PubMed Central also includes entity mark-up for the abstract text and a Bio Entities tab. In addition there is a “HAS_CHEMBL:y” tag that can identify those publications extracted by ChEMBL with the entities marked up. This is focused on medicinal chemistry rather than pharmacology papers but note the latter may cite the former.
Appendix V Patents
In terms of global drug discovery output these contain more medicinal chemistry data than papers (see PMID 24204758). However, compared to journals, they are more difficult to mine. While tips for this cannot be included here, anyone interested in patent searching the context of new or existing GtoPdb content is welcome to contact us. Note we routinely cross-check sources such as SureChEMBL in the case of new clinical candidates to see what expanded SAR data sets may be available.
New content and features: November 2014 update
The latest database version (2014.3) was released on 5th November and includes many content updates and new website enhancements. The current release now covers 7220 distinct ligands interacting at 2708 protein targets (see full statistics on our About page). This post describes all the changes:
Website
- Overview pages for target classes and superfamilies: while browsing the target family lists you can now click on the ‘Overview’ icon to access overviews, subfamily lists and further reading for target super families (Figure 1)
- Search term auto completion: As you type in search terms which match target or ligand names in the database these now come up as a list of suggestions (Figure 2). Clicking on a result links straight to the database page for that entity.
- Searches now work with Greek characters e.g. α,β.
- Removed option to choose whether to go to the concise or detailed view page in target search results. Results link directly to the detailed view if it exists, or to the concise view if it doesn’t.
- Data downloads: new peptide ligand CSV file with sequences and chemical modifications.
Figure 1 – Screenshot of the enzyme family list showing the new overview icon which links to superfamily overview pages.
Figure 2 – Screenshot of search term auto complete suggestions for target and ligand names
Target updates
GPCRs:
- Class Frizzled GPCRs
- Lysophospholipid (LPA) receptors
- Lysophospholipid (S1P) receptors
- Melanin-concentrating hormone receptors
- Melatonin receptors
- Neuropeptide FF/neuropeptide AF receptors
- Adenosine A<sub>2A</sub> receptor
- Complement C5a<sub>1</sub> receptor
- Complement C5a<sub>2</sub> receptor
- Dopamine D<sub>3</sub> receptor
VGICs:
- Inwardly rectifying potassium channels
- Voltage-gated calcium channels
- Selected members of the Two-P potassium channels
- Selected members of the Voltage-gated potassium channels
- Selected members of the Voltage-gated sodium channels
Enzymes:
- Kinases: curation of all phase III kinase inhibitors
- Proteases:
Ligands:
- Repurposing compounds from NCATS, AstraZeneca Open Innovation and other sources
- AZD compounds
- Repurposing
GuidetoPharmacology.org is the new home of IUPHAR-DB
Please note that the original iuphar-db.org site now redirects to guidetopharmacology.org. All target and ligand pages automatically redirect to the corresponding pages on the new site and we are working with other sites that point to us to update their links. Please update your bookmarks and start using the new site. Feel free to send us any comments – we are always looking for ways to improve our site!
Our upcoming release
As we approach our 2014.3 release in a few weeks time the curation team are busy adding and updating records. This includes a round of inputs an revisions from our protein family committees and a new FAQ. Just to remind you (as we hope we have also made clear in the past) we welcome suggestions of new content (that we may be able to squeeze in to this release), particularly from your own recent publications or “hot topics” you think are worthy of inclusion. In the first instance this can be to simply send a URL for the publication to this email address.
As you can see from the database content we use what can be termed primary data (e.g. an in vitro IC50) for new ligands and targets. However, we certainly consider secondary publications on new in vivo or clinical pharmacology (but try to send us the primary citations as well).
Keep an eye out for news of our release, and the new content and features over the coming weeks.
BPS Focused meeting: Exploiting the new pharmacology and application to drug discovery
A British Pharmacological Society (BPS) focused meeting in association with NC-IUPHAR will take place in Edinburgh in 2015, exploring new pharmacology and its application to drug discovery. This two-day event running from 20th-21st April will include talks by keynote speakers Sir Colin Dollery and Bill Catterall, and cover topics including new strategies for targeting calcium channels, tyrosine kinases, microRNAs, epigenetics, allosteric modulators and biased signalling.
The flyer for the event can be accessed via the link below, and includes contact details for the BPS and a list of speakers.
Exploiting the new pharmacology and application to drug discovery
NC-IUPHAR newsletter Summer 2014
The latest edition of our newsletter can be read at the link below:
NC-IUPHAR_Newsletter_Summer_2014
Special features covered in this issue include:
- A detailed profile of the PubChem database contributed by their lead scientist Evan Bolton, with whom we have worked in consultation with as we move forward with our ligand curation;
- “Out-and-about with the database team” – a summary of conferences and meetings attended by the team so far this year;
- Spotlight feature on our curation of approved drugs;
- “Pharmacology of human proteases”, contributed by Tony Turner, chair of the NC-IUPHAR committee for proteases and hydrolases;
- A tribute to NC-IUPHAR Vice Chair and Database Chair Prof. Tony Harmar, who sadly passed away earlier this year.
Regular features include:
- A message from NC-IUPHAR chairman Michael Spedding;
- An update from the database team on our work on the IUPHAR/BPS Guide to PHARMACOLOGY;
- ‘Student experiences’, this time with a piece contributed by our 2013 summer student Katerina Gospodinova;
- A report of our April 2014 NC-IUPHAR meeting
The newsletter is also available via our website and we have also created an archive of previous issues which can be viewed here.
WCP2014: Abstracts from NC-IUPHAR members
The 17th World Congress of Basic and Clinical Pharmacology (WCP2014) is now rapidly approaching and several NC-IUPHAR members will be attending the Congress. The followings talks will be presented on Tuesday 15th July 2014 15.30-17.00 in Track 6 of the programme: NC-IUPHAR and the IUPHAR/BPS Guide to PHARMACOLOGY.
From Sir Colin Dollery, Clinical Translational Pharmacology group:
NC-IUPHAR 1987-2014 and beyond
At the Sydney, Australia IUPHAR Congress in 1987, when I became President of IUPHAR, the Executive Committee decided to establish NC-IUPHAR, a committee on receptor nomenclature. At this time there was very little structural knowledge of receptors but a standardised nomenclature system was badly needed. The rapid ascent of NC-IUPHAR owed much to its first chair, Paul Vanhoutte’s energy, persuasive talents, and infectious enthusiasm. It was clear that a critical convergence was taking place between experimental and molecular pharmacology with a need to standardise nomenclature and provide an authoritative review of the evidence published on the properties of these molecules and the substances that interacted with them. A critical decision was to form over 60 expert committees with world wide membership to ensure a high quality of review. Since then there has been an exponential growth in activity, following the sequencing of the whole genome. Successive chairs, Bob Ruffolo and Michael Spedding, have contributing greatly to sustaining the quality and momentum of NC-IUPHAR as have those who have maintained the database. The volume and complexity of the data available continues to expand and the scope of the NC-IUPHAR-DB has expanded with it. Initially focused on GPCRs it has expanded to include ion channels, nuclear receptors, homo and heterodimers and current work on the kinome. NC-IUPHAR has an h-index of >60 and has a freely available database on receptors which will be described fully in this symposium. As the volume of BIG Data explodes the need for critical curation to maintain high standards of accuracy in documentation of targets and ligands in pharmacology and clinical pharmacology will grow too.
From Michael Spedding, NC-IUPHAR chairperson:
How two decades of NC-IUPHAR have helped resolve controversy in drug discovery
NC-IUPHAR (the nomenclature committee of IUPHAR) is engaged in a major task: How can we define all the main receptor and drug targets coded by the human genome, and put them in a database (guidetopharmacology.org) freely accessible to all? Even more important, link them to therapeutics and pharmacological target validation? The immense recent growth of knowledge about drug targets, with their crystal structures, has been a huge help to drug discovery and while IUPHAR classifications are regularly used, we are able to be proactive including targets, and contacting experts. We have 80 subcomittees of >700 pharmacologists contributing in their relevant fields, organised by our 5 curators, who are financed by the IUPHAR, the Wellcome trust, the BPS and our industrial sponsors. We have 2500 proteins in the database, with >6000 ligands, ~600 approved drugs. With the vast increase in knowledge, why is drug discovery not more successful? Perhaps because the increase in knowledge has been matched by the increase in variables affecting drug-receptor interactions. This is why we try and include data on how the new ‘knowledge frontiers’ of alternative spicing, non-coding RNAs, epigenetics, allostery, and biased signalling may affect drug discovery and development. An example : Doriano Fabbro, with Elena Faccenda as curator, has produced a complete database of kinases – with their pharmacology, thanks also to Novartis, Discoverx, Millipore and Reaction Biology providing data. This is an expert-based system, based on validated data, freely available to all, in our publications or on http://www.guidetopharmacology.org. However, these variables also lead to controversy and disagreement, sometimes fierce, between experts. NC-IUPHAR is the arbiter, and sometimes the target, of occasionally vivid controversy. Some examples will be given! But this means that we are relevant. NC-IUPHAR is open to all pharmacologists!
From Adam Pawson, Senior Database Curator:
IUPHAR-DB, GRAC and the IUPHAR/BPS Guide to PHARMACOLOGY
The International Union of Basic and Clinical Pharmacology/British Pharmacological Society (IUPHAR/BPS) Guide to PHARMACOLOGY (http://www.guidetopharmacology.org) is a new open access resource providing pharmacological, chemical, genetic, functional and pathophysiological data on the targets of approved and experimental drugs. Created under the auspices of the IUPHAR and the BPS, the portal provides concise, peer-reviewed overviews of the key properties of a wide range of established and potential drug targets, with in-depth information for a subset of important targets. The resource is the result of curation and integration of data from two well-established resources: (1) the IUPHAR Database [IUPHAR-DB; the original database of the IUPHAR Committee on Receptor Nomenclature and Drug Classification (NC-IUPHAR) and driving force behind the development of the Guide to PHARMACOLOGY] and; (2) the published BPS ‘Guide to Receptors and Channels’ (GRAC) compendium. The data are derived from a global network of expert contributors, and the information is extensively linked to relevant databases, including ChEMBL, DrugBank, Ensembl, PubChem, UniProt and PubMed. Each of the ∼6000 small molecule and peptide ligands is annotated with manually curated 2D chemical structures or amino acid sequences, nomenclature and database links. Future expansion of the resource will complete the coverage of all the targets of currently approved drugs and future candidate targets, alongside educational resources to guide scientists and students in pharmacological principles and techniques. GRAC has now been superseded by ‘The Concise Guide to PHARMACOLOGY’, a biennial snapshot of the overviews of the key properties of each target family, intended to be a quick desktop reference guide.
From Chris Southan, Database Curator and Chairperson of the NC-IUPHAR Drugs and Targets Annotation Subcommittee
Analysing the drug targets in the human genome
Discerning the molecular mechanisms of action (mmoa) for drugs treating human diseases is crucially important. This talk will provide an overview of target numbers in IUPHAR/BPS Guide to PHARMACOLOGY, compare these to other sources and consider the wider implications for drug discovery. We have developed stringent mapping criteria for primary targets (i.e. identifying those direct protein interactions mechanistically causative for therapeutic efficacy). This includes inter-source corroboration by intersecting multiple drug sources inside PubChem to produce consensus structure sets. The analogous approach is used to intersect published target lists and database subsets at the UniProtKB/Swiss-Prot identity level. Our cumulative curation results reveal that structure representation differences, data provenance and variability of assay results, are major issues for experimental pharmacology and global database quality. While our activity mappings encompass some polypharmacology (e.g. dual inhibitors and kinase panel screens) our strategic choice is to annotate minimal rather than maximal target sets. The consequent increased precision gives our database high utility for data mining, linking and cross-referencing. Our small-molecule figures are currently converging to ~200 human protein primary targets for ~1000 consensus chemical structures of approved drugs. Target lists from other sources are typically larger and show a degree of discordance. Comparative analysis of these lists by their UniProt ID content and Gene Ontology distributions suggests differences in curatorial selection are the main cause of divergence. The global target landscape thus shows paradoxical trends. On the one hand, cumulative drug research output and recent expansions (e.g. epigenetic targets and orphan diseases) have pushed bioactive compounds from papers or patents to above 2 million and chemically modulatable human proteins above 1500 (PMID:24204758). On the other hand, reports of Phase II clinical efficacy failure, with implicit target de-validation, are frequent. In addition, our assessment of drug approval data from 2009 to 2013 indicates new targets (i.e. first-in-class mmoas) are so low as to threaten the sustainability of the pharmaceutical industry. Causes and consequences of these paradoxes, along with utilities for minimal and maximal druggable genomes, will be discussed.
From Simon Maxwell, Educational Site Project Leader
The IUPHAR/ASPET Pharmacology Education Project
Pharmacology has been the science at the heart of many of the advances in healthcare that we now take for granted. Those advances have depended on expertise in all areas from basic pharmacology through translational research to prescribing and monitoring the use of medicines by patients. Each step has required individuals with a clear grasp of the fundamental principles of pharmacology and clinical pharmacology supported by knowledge of the actions of specific drugs. For many scientists, and in many academic institutions, there has been an erosion of the visibility and profile of the pharmacological sciences as we move progressively into an era of inter-disciplinarity. The IUPHAR/ASPET Pharmacology Education Project seeks to bridge this gap and promote learning in pharmacology to a worldwide audience. The specific aims of the project are to: (i) produce a simple, attractive easily searchable resource that will support students of the pharmacological and other biomedical sciences, as well as health professions students including medicine, nursing, pharmacy and others; (ii) make high quality resources available to support pharmacology educators; and (iii) service the needs of students and teachers in resource-poor countries or where pharmacology is less well developed. The project has been supported for the first two years by a generous grant from ASPET and will be led by an international Editorial Board bringing together individuals with demonstrable commitment to pharmacology education. That group will welcome the input of colleagues from around the world as it brings together links to existing resources and gradually creates new ones. The content will be divided broadly into four sections: pharmacology, clinical pharmacology, drugs and therapeutics. Each section will be divided into modules, which will have associated topics to which learning resources will be attached (e.g. web-links, articles, video and audio files, PowerPoint). The longer-term aspiration would be that each topic would have an associated summary text generated by the editors. In addition to this simple structure there might be additional content including a formulary, glossary, and discussion board.
Additionally, Chris will be presenting a poster entitled ‘Will the real drugs please stand up? with the following abstract:
BACKGROUND: A comparison of database subsets of approved drugs in 2009 recorded only 807 exact structures in-common (PMID:20298516). Factors contributing to low overlap included semantic naming inconsistencies, ambiguity in structure representation and the fact that neither regulatory bodies nor pharmaceutical companies directly verify public electronic chemical database records. This work is a current comparison of drug sources inside PubChem.
METHODS: We selected submitters that nominally included small-molecule drug collections and International Non-proprietary names (INNs) and/or US approved names (USANS). Unions, intersects and differences were derived by using the Entrez query history interface to perform Boolean operations on retrieved sets. Additional filters were explored, including salt-stripping by selecting a covalent unit count of 1.
RESULTS: DrugBank 3.0 declares 1,541 small-molecule drugs and the term “approved” returned 1,424 substances (SIDs) in PubChem. These collapse to 1,392 compounds (CIDs), and removal of mixtures reduces to 1,325. The Therapeutic Target Database (TTD) declares 1,540 approved drugs on their website. The CID overlap with the DrugBank 1,325 was 1,108, and the equivalent figure for ChEMBL_17 was 1,141. The three-way consensus (from the DrugBank starting point) was 1,003. The term INN retrieves 7,916 CIDS, reducing to 7,180 single-components. USAN brings back 5,494 of which only 3,204 are single-component (i.e. more salt forms are designated as USANs). Of the 1,108 3-way set, 927 have an INN or USAN. The “same connectivity” query indicates, on average, each of the 927 have nearly 20 canonically-related CIDs. Issues associated with these metrics will be outlined and, depending on new source releases, the numbers will be updated.
CONCLUSIONS: A surprising degree of non-overlap persists in drug structures. Our results are not a criticism of the valuable sources but further analysis is needed of the multiplicity of structural representations and fuzzy naming of essentially the same canonical drugs inside PubChem. This important issue in cheminformatics extends beyond the INNs to all pharmacologically active structures. It also rationalises our IUPHAR/BPS Guide to PHARMACOLOGY strategic choice of focusing on consensus sets for curation. This work indicates definitive drug lists will remain elusive until there is more collective engagement for provenance, standardisation and cross-mapping.
This poster (no.529) will be displayed on Monday 14th and Tuesday 15th July but is also available to view on SlideShare.
Case studies in chemical curation III: complexes and component mixtures
The issue of specified mixtures that include pharmacologically active chemical structures is complex and curatorially challenging, as described by this presentation on the topic.
This post will briefly cover our handing of salts. A substance in an approved drug preparation may not only be specified as a salt in national regulatory documentation but also in the physical molecular form within the tablet itself. A well known example is atorvastatin calcium (or more strictly, from the FDA insert label, hemi-calcium trihydrate, CID 656846). For many reasons (primarily our MMOA and activity-mapping focus) we have opted for the stringency of representing “parents” (i.e. not salts) as primary ligands, as can be seen in our ligand entry.
Database metrics can be used to illustrate the complexities in drug data sets and also supports our approach to simplifying the relationship mappings. PubChem (June 2014) contains 8072 CIDs with an INN and 5636 with a USAN (the caveat is these tags are from secondary sources, not the authorities themselves). Of these, INN are 91% singleton “parents” but only 58% for USANs. While the pharmaceutical importance of salt forms is clear, to enhance the clarity of our ligand entries we usually align them to the parent INN structures. However, we may include links to pharmacologically-relevant salt forms. If you want to see all mixtures (salts and/or combinations) associated with a ligand you can click on the “Related substances, Mixture” tag in the PubChem CID (the caveat is that for well established drugs these can be large “overkill” lists from patent-only specified salts or mixtures).
One of the issues we face for in vitro activity mapping is that bioassay data is often split between database records of not only parents but also different salt forms specified in different extractions of experimental descriptions. We make our judgments based on inspecting the original papers but where active compounds are extensively diluted into assay buffers, our inter-publication normalisation to parent is a justifiable simplification. Those wishing to perform similar experiments would obviously need to check the reference, for example which salt a commercial reagent source may have specified (but sometimes not).
Contributed by Chris Southan and Helen Benson
Guide to PHARMACOLOGY out and about: Spring 2014 conferences and meetings
In April one of our curators, Helen Benson, represented the team at the Seventh International Biocuration Conference at the University of Toronto, and presented a poster outlining our approved drug and clinical target curation. This visit followed the previous year’s meeting of the International Society for Biocuration in Cambridge, UK, attended by both Helen and database developer Joanna Sharman.
Over the 4 days of the conference, the programme of talks and workshops covered many themes relevant to our work, as well as discussing the challenges facing biocurators today. These challenges include how curatorial work will evolve in an age of big data, and the role of crowdsourcing and text mining in managing the volume of biological data to be included in online databases. Keynote speakers Suzanna Lewis and Lincoln Stein discussed these themes with their respective talks on optimisation of curation and whether Big Data will crush curation.
One of the main themes from the talks and the workshops was the role and implementation of clinical and anatomical ontologies to databases. Additionally, several talks discussed data standards and controlled vocabularies, with particular regard to pathway curation. This issue is particularly pertinent to us as we now include enzyme pathways in our database.
Our team of curators has now expanded to 4, and our pipeline for ligand and target curation is constantly evolving to achieve optimum methods. It was therefore invaluable for us to learn more about the projects of other research groups, and to hear about which areas have become key focus points for the field. As always, it was a pleasure to meet others working in the field and we look forward to attending this event in the future.
Our database developer, Joanna Sharman, attended Edinburgh Neuroscience Day, an annual day of talks and poster presentations involving over 300 researchers with an interest in neuroscience.
Joanna also attended and presented a poster at the Barcelona GPCR Spring Conference organised by the GLISTEN network. This included presenting and discussing proposals for collaborations at the GPCRDB Satellite Meeting.
Joanna was also invited to attend a workshop on Computational Challenges in Data Citation at the University of Pennsylvania, Philadelphia, which brought together three groups of people (Computer Scientists, Information Scientists and Data Scientists) to explore the technical challenges and research opportunities posed by the increasing demand to generate citations for large, complex datasets.
Chemical curator Chris Southan started his 2014 representations with an invited visit to the April 29th – May 1st BioIT World, Boston, as co-organiser and presenter at a Workshop entitled “A Bar Code for Chemical Structures:Using the InChI to Transform Connectivity between Chemistry, Biology, Biomedicine and Drug Discovery”.
His individual workshop presentation was entitled “Transformative Utility of InChIKey Searching in the Mother of all Databases”.
A poster related to the database was also presented on “Will the real drugs please stand up?”.
In May 6th Chris had the privilege of being one of the opponents in a (successful) PhD examination for The Faculty of Pharmaceutical Sciences, University of Copenhagen. The trip included a presentation to the department and the GPCRDB team entitled “Will the real drugs and targets please stand up ? Evolving consensus-based curatorial strategies”.
On June 1-5th Chris attended the 10th International Conference on Chemical Structures. His oral presentation was entitled “Will the real drug targets please stand up?”.
As an alumnus from his work on the ELIXIR project in 2008/9 Chris attended the June 12th EMBL-EBI 20th Anniversary celebrations. He also took the opportunity to visit the EBI and Cambridge on the next day for discussions with a range of collaborators including UniProt, MEROPS, ChEMBL and NextMove Software.
Database Principal Investigator Jamie Davies attended the SULSA Synthetic Biology Meeting, in Edinburgh on 10th June and presented our poster “Exploiting Edinburgh’s Guide to PHARMACOLOGY database as a source of protein design information for synthetic biology”.
New features on the Guide to PHARMACOLOGY: June 2014
Our latest database update took place on Monday 16th June and this post summarises the new content and features on the site.
Website demo
Our ‘Help’ pages now include a ‘walkthrough’ demo of the website produced by Prof. Tony Harmar prior to his retirement. This resource complements the tutorial already available via our ‘Resources’ menu and we especially hope our new users will find it valuable for getting to grips with the site.
Target updates
Our enzyme target class has been updated to include additional chromatin-modifying enzymes from several recent review articles, while our ‘other proteins targets’ class now includes bromodomain-containing proteins, also curated from recent reviews. Many of these target pages have also now been updated to include interaction data for the ligands which modify these targets. For the GPCR target class, the dopamine D1 and somatostatin sst3 and sst5 receptor pages have been updated.
Ligand updates
We now include a separate tab on our ligand list for labelled ligands. This includes radiolabelled ligands, unstable isotopes, fluorescent tags and small chemical entities.
Each of the tabs on our ligand list is now annotated with our ‘approved drug’ symbol, indicating approved drug ligands within each class.
Following a quality-control check on our ligand entries in consultation with PubChem, many of our ligand pages have been further annotated with CID-crosslinks and contextual comments. Our series of blog posts on chemical curation will discuss some of the interesting case studies we encountered during this exercise.
A summary of the total number of targets and ligand now in the database is available on our ‘About’ page