GtoPdb and its precursor IUPHAR-DB have been capturing the structures of pharmacologically relevant ligands since 2005. The fig.1. snapshot below shows the approved drug section of our eight-category ligand classification
As an active collaboration with the PubChem team, we have submitted our ligand records for every GtoPdb release since 2012. For the current release of 2016.4 the query (“IUPHAR/BPS Guide to PHARMACOLOGY”[SourceName]) retrieves 8674 Substance Identifiers (SIDs) and 6565 Compound Identifiers (CIDs). The excess of 2109 SIDs is accounted for by antibodies, small proteins and large peptides that cannot form CIDs. At just over 92 million CIDs from 473 sources, a range of property filters and full Boolean operations for combining query sets, PubChem provides an opportunity to “slice and dice” our ligand set in detailed, comparative and informative ways. A set of results is shown below.
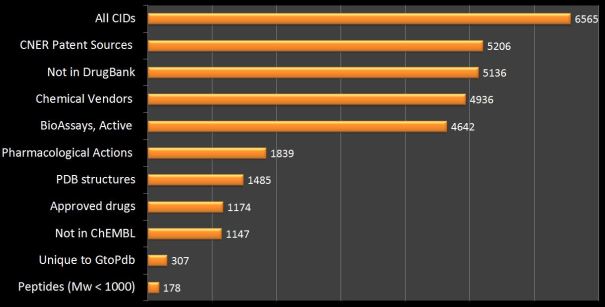
The utilities of these intersects are outlined below (in order of counts):
- CNER refers to “Chemical Named Entity Recognition” for the automated extraction of chemistry from patents by sources submitting to PubChem (of which SureChEMBL is the largest at 16.3 million). This means that users can track-back most of our ligands to early patent filings that can often include more SAR than eventually appeared in the papers.
- Our low overlap with DrugBank indicates both sources are complementary in bioactive compound selection (i.e. the OR union is 12605)
- The possibility of sourcing purchasable compounds is important for experimental pharmacologists. From the 64 million vendor structures in PubChem we have nearly an 80% overlap and similarity searches may pick up analogues where there is no exact match.
- The “BioAssay active” tag overlaps extensively with ChEMBL entries but users can check for a range of activities for a ligand that maybe additional to the values we have extracted from selected papers.
- The MeSH term “pharmacological action” is useful but our impression is that NLM is falling behind in the PubChem indexing of this term.
- PDB ligand structures are valued database cross-references for many reasons.
- We have introduced a new feature that allows users to retrieve just our 1291 approved drug SID entries (Query “approved[Comment] AND “IUPHAR/BPS Guide to PHARMACOLOGY”[SourceName]”). The “PubChem Same Compound” select then generates 1174 small-molecule CIDs. This facilitates different types of comparative analysis between drug lists.
- As expected, our overlap with ChEMBL structures is high but we have captured 1147 structures not in this source, mainly due to different journal capture and shorter release cycles.
- The selection “unique to GtoPdb” indicates those CIDs where we are the only source in the whole of PubChem. These are predominantly novel structures we have extracted from papers but in some cases we have selected a different structure from other sources.
- There may be interest in which pharmacologically active peptides we have CIDs for. A simple Mw-cut isolates 178 entries
In regard to 7) a snapshot from our list of approved drugs is shown below




Leave a comment