This post covers three recent publications with a common theme and whose authors are collaborators with GtoPdb, thus making them as a trio particularly suitable for combined review. These are; Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors [1], Novel approaches leading towards peptide GPCR de-orphanisation [2] , and Advances in therapeutic peptides targeting G protein-coupled receptors [3]. As expected, these are all clearly written, contain valuable detail and each includes its own introduction. Therefore this piece does not need to go over these works per se but simply point out their coverage, domain of application, along with brief explanations of entity linking and/or external look-up options.
For context, it should be pointed out that historical success in de-orphanising GPCRs (i.e. reproducibly pairing them with physiologically-relevant endogenous ligands) has been difficult and slow. This remains so, despite decades of intense effort from not only all major pharmaceutical companies but also academic groups utilising various flavours of de-orphanistaion screening approaches and technologies. This was pursued in parallel with cloning (and patenting) the complete repertoire of human GPCRs as they slowly came into view. Initially done via mining millions of expressed sequence tag (EST) transcripts in the mid 90’s, this later shifted focus to genomic DNA surfacing between appropriately 1998 and 2000. The modest progress even after the completion of the draft human genome can be discerned from the table below compiled in 2000 [4].
Progress ~ 2000- 2018 is outlined both in Concise Guide 2019/20 : G Protein-Coupled Receptors (PMID: 31710717) [5] and in the Guide to Pharmacology latest parings list .
As the first of this trio of papers, Foster et al. [1] have significantly advanced the de-orphanisation field in 2019 by integrating computational and experimental approaches for peptide-GPCR pairings. They began with comparative sequence and structural analyses to gain biological insights into the human peptide-receptor signalling landscape. They were then able to leverage these features to mine candidate peptide ligands from the entire human genome, arranged for these to be synthesised and then tested for activity in a broad range of pairing assays that included known ligands as positive controls. The impressive scope of results is indicated in their Fig 7 (below).
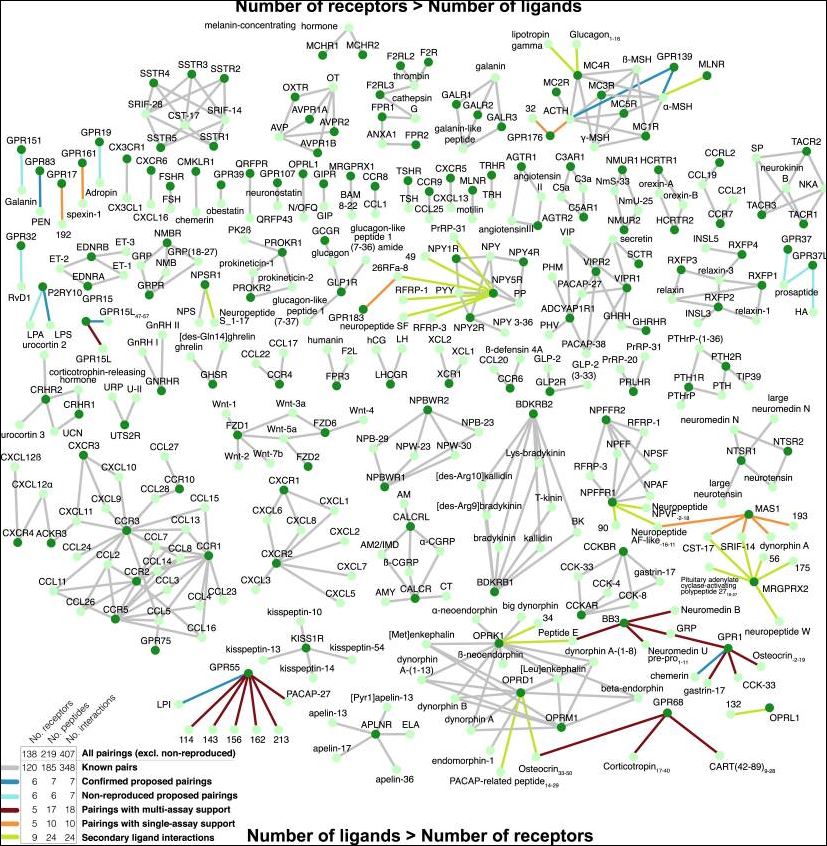
In summary, the 53 peptides with validated receptor-dependent responses first reported in this paper, has expanded the known human peptidergic signaling network from 348 to 407 interactions. It is also important to note that they were unable to reproduce no less than six reported parings in the literature from 2004 to 2016 (i.e. those de-orphanisations could not be verified).
In collaboration with the Copenhagen crew, this paper was curated by the GtoPdb team according to their target mapping stringency and potency thresholds. Consequently, 11 new peptides have been annotated with the quantitative ligand binding data reported for the newly de-orphanised receptors. These are shown below with their PubChem Substance Links (SIDs) for which GtoPdb (so far) has becomes the only source of these compound structures (CIDs) from over 730 submitters.
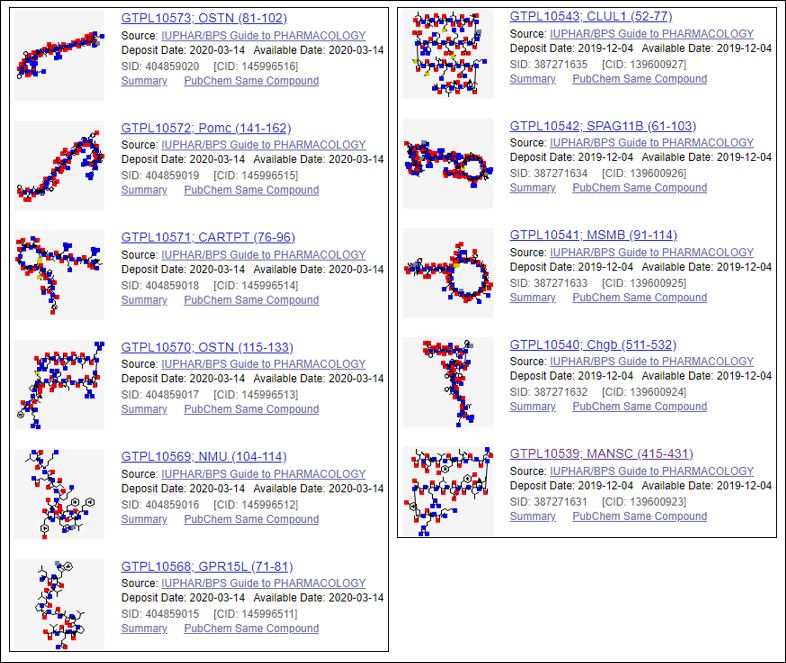 In PubChem these can be accessed by scrolling down to “Related information” on the lower right hand facet of PMID: 31675498 and clicking on “PubChem Substance”. Each entry has a pointer back to GtoPdb from the SID and the nomenclature of the peptides matches that used in the paper. However, users wanting to select these entries from within GtoPdb can go to Ligand Search Tools > Search for data by literature reference > Enter PubMed Id: 31675498 > Select field to search: PubMed ID. This brings back 16 entries since it includes the 11 ligands above plus the five receptors for which binding data was curated (n.b. these instructions apply to the old PubMed interface, so some of this may change for the new interface). While the version of the article in Cell journal itself has no entity outlinks (except for references) the PubMed Central version has automatically linked the PDB IDs. All the GPCR protein/gene names should retrieve in GtoPdb or of course GPCRdb
In PubChem these can be accessed by scrolling down to “Related information” on the lower right hand facet of PMID: 31675498 and clicking on “PubChem Substance”. Each entry has a pointer back to GtoPdb from the SID and the nomenclature of the peptides matches that used in the paper. However, users wanting to select these entries from within GtoPdb can go to Ligand Search Tools > Search for data by literature reference > Enter PubMed Id: 31675498 > Select field to search: PubMed ID. This brings back 16 entries since it includes the 11 ligands above plus the five receptors for which binding data was curated (n.b. these instructions apply to the old PubMed interface, so some of this may change for the new interface). While the version of the article in Cell journal itself has no entity outlinks (except for references) the PubMed Central version has automatically linked the PDB IDs. All the GPCR protein/gene names should retrieve in GtoPdb or of course GPCRdb
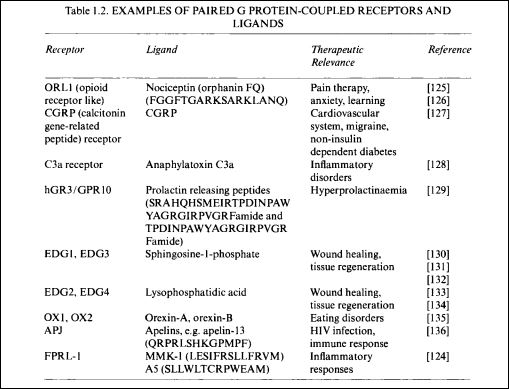
The second article in the trio is a review by Hauser et al [2]. This covers some of the same ground as above but includes an assessment of contemporary methodologies of de‐orphanisation. The informative Figure 1, summarising extensive data mining, is shown below.
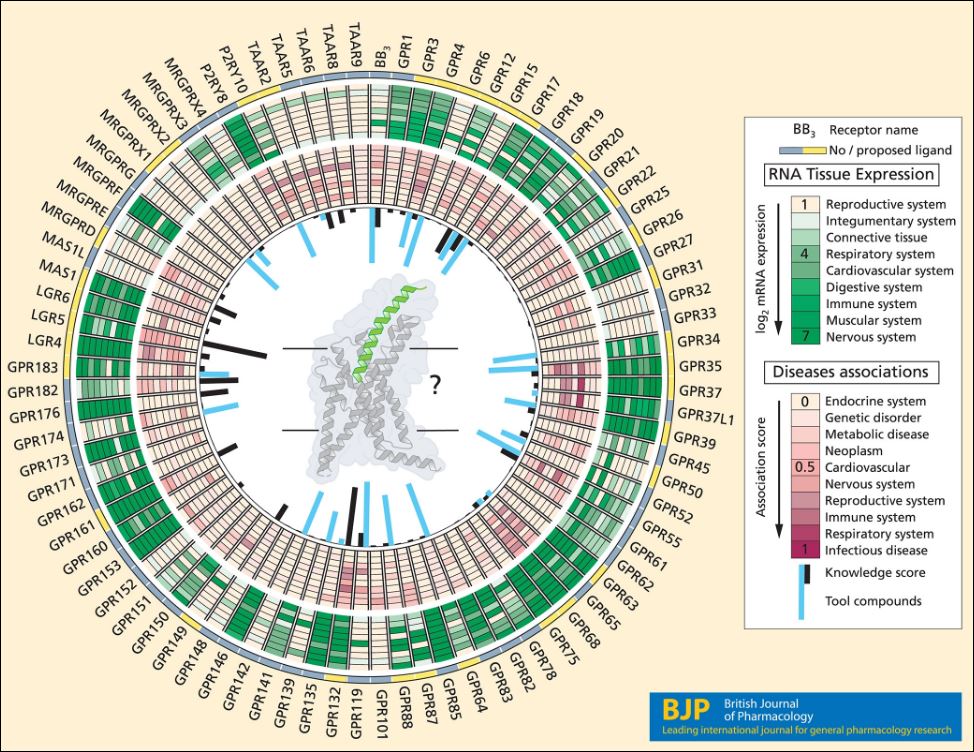
In addition to indicating the ligand status of the GPCRs the circles extend to clinical relevance, disease therapeutic potential, publication density and tool compounds from ChEMBL. Note that while ChEMBL links to compounds with a wide range of potentcies GtoPdb has a more stringent annotation of smaller number of probe candidates including very recent publications (e.g. these 11 ligands above are not in ChEMBL26).
One of the advantages of publishing in BJP is the provision of author-specified out-links to GtoPdb ligands and targets (see PMID 30087946). These have been duly added to the PDF for all GPCRs mentioned in the text as well as any ligands that were already in GtoPdb before this review. Examples for five links are shown below.

Clicking on “GPR15” above thus takes us the full annotation for that receptor, including the agonist ligand records shown below.
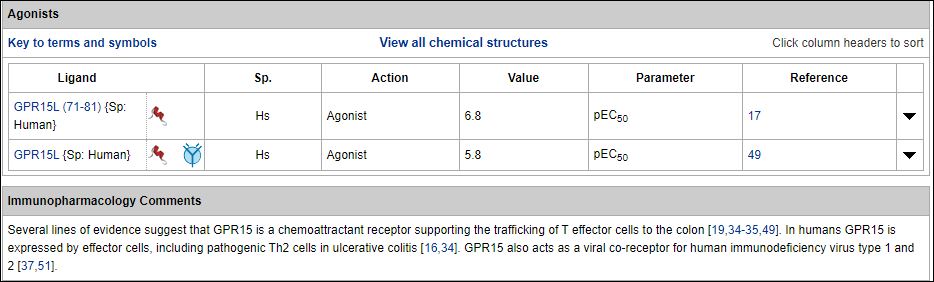
Thus GPR15L (71-81) corresponds to SID 404859015. The format of Table 1 unfortunately precluded the addition of the live-links for the new ligands.
The last of this trio is an in-depth review by Davenport et. al. [3] that encompasses translation aspects of peptides that modulate GPCRs into clinically effective drugs. This may even eventually extent to analogues of the peptides from the first two papers perhaps even related to new de-orphanisations. As detailed in this review therapeutic peptides have recently been undergoing a development technology renaissance via half- life extension, stapling and resistance to proteolysis. These measures improve pharmacokinetics and oral bioavailability that have hitherto been much less favourable for peptidic structures in comparison to conventional small-molecule drug candidates. An overview of these peptide approvals and advanced candidates is taken from their Figure 1.
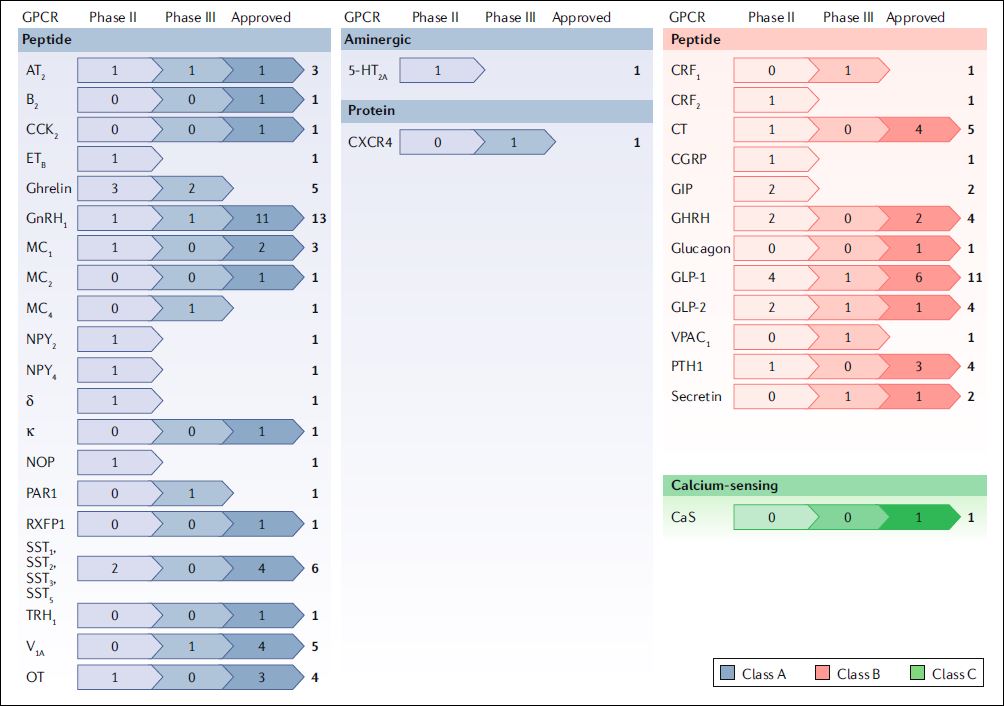
This comprehensive review goes on to describe in detail 26 synthetic peptides approved for clinical use (20 agonists and 6 competitive antagonists) targeting eight class A receptor families. It also includes a useful structural perspectives not only on peptide binding sites but also addresses the frequently overlooked aspects associated with unnatural amino acids and chemical modifications.
It seems somewhat anachronistic that the Nat Rev Drug Discov PDFs are entity link-free zones (although this FAIR-gap has been pointed out to them). Notwithstanding, the GPCR names should retrieve cleanly from GtoPdb and GPCRdb. Most of the peptide names in the review should also retrieve their ligand entries from GtoPdb but these will eventually be cross-checked both for the resolution of equivocalities and addition of new development candidates with referenced binding data.
Constituting a well-referenced overview of the field, these three papers also represent “good news” for peptides and their analogues. Notwithstanding, this slide set outlines some of the challenges for curating peptides and searching them in databases.
**********************************
Comments by Chris Southan, Fellow of the University Edinburgh, Owner of TW2Informatics , Chair of NC-IUPHAR Subcommitees for Proteases and Drug Targets and Chemistry (DRUTACS), ORCID 0000-0001-9580-0446. As declaration of interest (but not conflict!) Chris was pleased to be a co-supervisor for Alex Hauser’s PhD work on Computational Receptor Biology
[1] Discovery of Human Signaling Systems: Pairing Peptides to G Protein-Coupled Receptors. Foster SR, Hauser AS, Vedel L, Strachan RT, Huang XP, Gavin AC, Shah SD, Nayak AP, Haugaard-Kedström LM, Penn RB, Roth BL, Bräuner-Osborne H, Gloriam DE. Cell. 2019 Oct 31;179(4):895-908.e21. doi: 10.1016/j.cell.2019.10.010. PMID: 31675498.
[2] Novel approaches leading towards peptide GPCR de-orphanisation. Hauser AS, Gloriam DE, Bräuner-Osborne H, Foster SR. Br J Pharmacol. 2020 Mar;177(5):961-968. doi: 10.1111/bph.14950. Epub 2020 Feb 3. PMID: 31863461.
[3] Advances in therapeutic peptides targeting G protein-coupled receptors. Anthony P. Davenport AP, Scully CCG , de Graaf C, Brown, AJH, Maguire JJ. Nat Rev Drug Discov (2020). https://doi.org/10.1038/s41573-020-0062-z.
[4] The impact of genomics on drug discovery. Beeley LJ, Duckworth DM, Southan C. Prog Med Chem. 2000;37:1-43. PMID: 10845246.
[5] The Concise Guide to PHARMACOLOGY 2019/20 : G Protein-Coupled Receptors. Alexander SPH, Christopoulos A, Davenport AP, Kelly E, Mathie A, Peters JA, Veale EL, Armstrong JF, Faccenda E, Harding SD, Pawson AJ, Sharman JL, Southan C, Davies JA; CGTP Collaborators. Br J Pharmacol. 2019 Dec;176 Suppl 1:S21-S141. doi: 10.1111/bph.14748. PMID: 31710717.






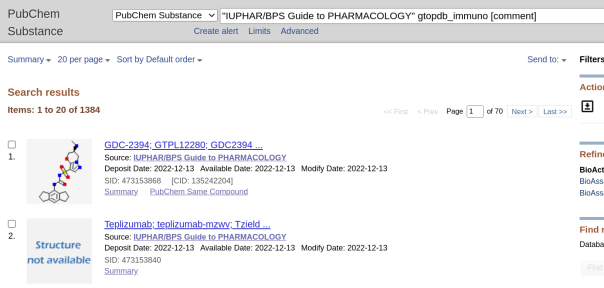









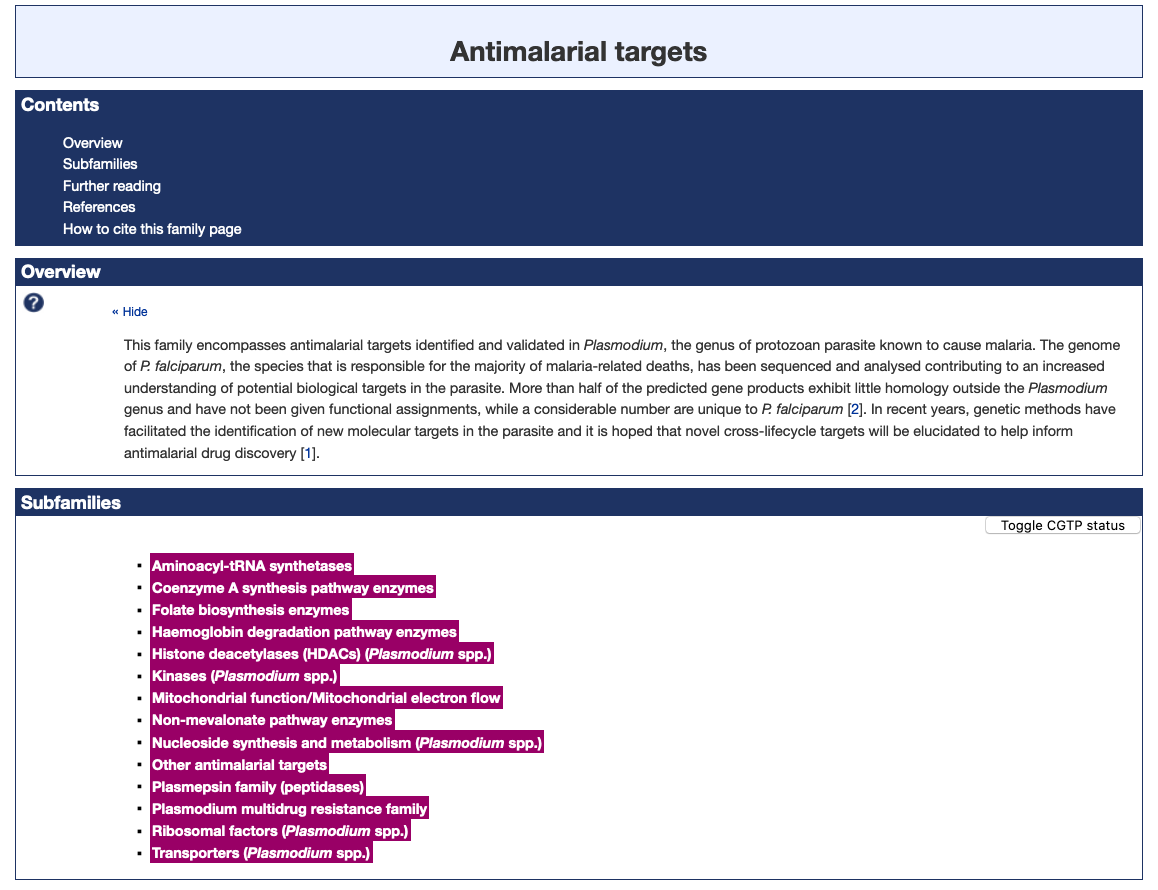 Figure 1. The Antimalarial targets family page illustrating the new subfamily classification (highlighted in magenta).
Figure 1. The Antimalarial targets family page illustrating the new subfamily classification (highlighted in magenta).






 In
In