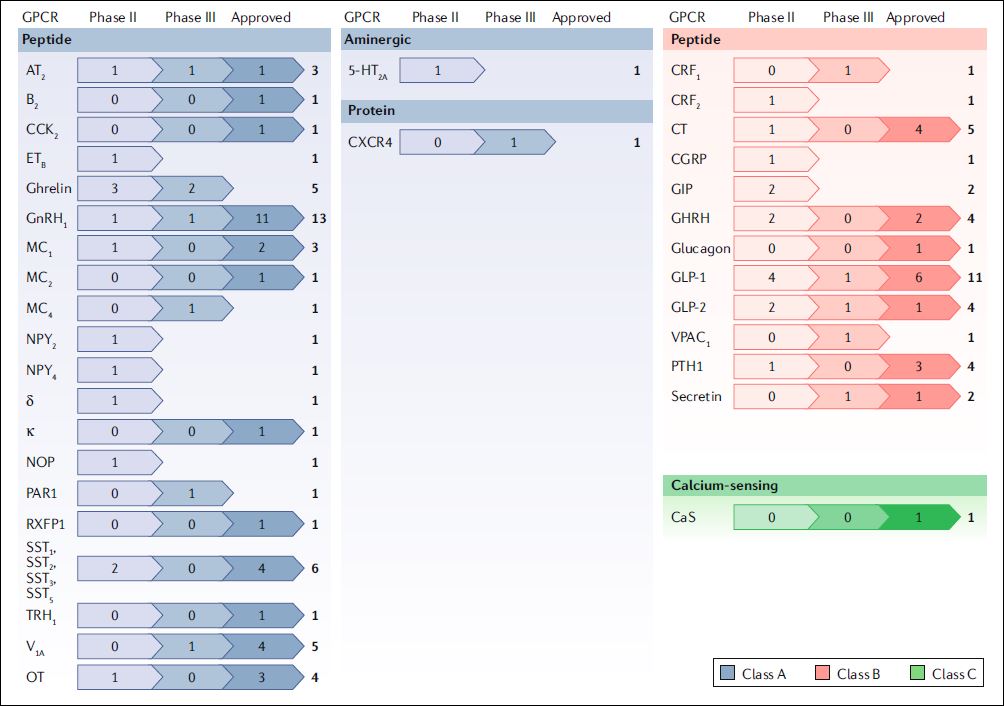
A new release, version 2024.1, has been made of the IUPHAR/BPS Guide to Pharmacology database. Released on 26th March 2024 this is the first release of the year. The following blog post gives details of the key content updates and website changes. GtoPdb now contains:
- 3,067 human targets, 1,732 of which have curated quantitative ligand interactions.
- 12,590 ligands, 9,204 of which have curated quantitative target interactions.
- 1,981 approved drugs, 1,128 with curated quantitative interactions.
- Clinical use summaries for over 3,704 ligands of which 1,971 are approved drugs.
- A total of 20,789 curated binding constants
- Data curated from over 45,000 references
Curation Update
Targets
11 new protein targets have been added to the GtoPdb since our last update. Ten of these are enzymes, with SIGLEC15 being the only new non-enzyme protein. PPP1CA, PPP2CA and ZDHHC3 were added as part of our natural products project. The remainder were added when new ligands for the targets were identified from literature searches
| TID | Family | Gene | Name | Comment |
| 3263 | Protein phosphatase catalytic subunits | PPP2CA | protein phosphatase 2 catalytic subunit alpha | Molecular target of natural marine toxins (okadaic acid, microcystin-LR, calyculin A) |
| 3264 | Protein phosphatase catalytic subunits | PPP1CA | protein phosphatase 1 catalytic subunit alpha | Molecular target of natural marine toxins (okadaic acid,
microcystin-LR, calyculin A) |
| 3265 | SIGLECs (conserved) | SIGLEC15 | sialic acid binding Ig like lectin 15 | Anti-SIGLEC15 mAb demonstrated experimental anti-tumour potential |
| 3266 | Mono-ADP-ribosylating PARPs | TIPARP | TCDD inducible poly(ADP-ribose) polymerase | Inhibitors are proposed to promote immunomodulatory and antineoplastic actions (atamparib, (S)-XY-05) |
| 3267 | Mono-ADP-ribosylating PARPs | PARP10 | poly(ADP-ribose) polymerase family member 10 | Dual PARP10/PARP15 inhibition might be beneficial in cancer treatment (see compound 8a [PMID: 35500474]) |
| 3268 | Mono-ADP-ribosylating PARPs | PARP11 | poly(ADP-ribose) polymerase family member 11 | The PARP11 inhibitor ITK7 is curated |
| 3269 | Mono-ADP-ribosylating PARPs | PARP14 | poly(ADP-ribose) polymerase family member 14 | Involved in the immune response and lymphocyte physiology; highly expressed in aggressive B cell lymphoma. RBN012759 used to explore anti-tumour potential of PARP14 inhibition. |
| 3270 | Mono-ADP-ribosylating PARPs | PARP15 | poly(ADP-ribose) polymerase family member 15 | Dual PARP10/PARP15 inhibition might be beneficial in cancer treatment (see compound 8a [PMID: 35500474]) |
| 3271 | 2.3.1.- Acyltransferases | ZDHHC3 | zinc finger DHHC-type palmitoyltransferase 3 | benzosceptrin C promotes T cell-mediated anti-tumour effect in vitro and in vivo; proposed as an alternative to anti-PD-L1 or anti-PD-1 immunotherapies |
| 3272 | S1: Chymotrypsin | CTRB1 | chymotrypsinogen B1 | Experimental inhibitor beta ph61 is curated |
| 3273 | AAA ATPases | CLPP | caseinolytic mitochondrial matrix peptidase proteolytic subunit | Validated target for the development of anti-tumour drugs; activator compound 16z [PMID: 35609303] and PAM ONC201 curated |
Ligands
Release of the newest proposed INN list (PL130) in February provided the opportunity to curate new nominally clinically-relevant entities. There were 414 INNs in this list. So far we have curated 27 kinase inhibitors from PL130, and where possible we have endeavoured to match the INNs to research codes in disclosed clinical pipelines, primary literature sources and clinical trials. We hope that this depth of curation provides additional insight and information for users of the GtoPdb. Occasionally the INNs match ligand structures that we have previously curated from other sources. Of note in PL130 was the inclusion of INNs for PROTAC type degrader molecules (zomiradomide, lirodegimod), and a kallikrein B1 gene editing agent (lonvoguran), both of which are relatively new classes of clinical interventions. We will continue to analyse this list to see if we can identify either new drug targets, or new pharmacological modalities for existing targets.
Drug approvals
Since the beginning of 2024 the FDA have approved 7 new drugs: as can be seen in the table below all but two of these are curated in the GtoPdb.
| Ligand ID | INN | Indication |
| n/i | berdazimer | To treat molluscum contagiosum |
| 10772, 12188 | cefepime, enmetazobactam | To treat complicated urinary tract infections |
| n/i | letibotulinumtoxinA-wlbg | To temporarily improve the appearance of moderate-to-severe glabellar lines |
| 9592 | tislelizumab-jsgr | To treat unresectable or metastatic esophageal squamous cell carcinoma |
| 12026 | resmetirom | To treat noncirrhotic non-alcoholic steatohepatitis with moderate to advanced liver scarring |
| 10070 | aprocitentan | To treat hypertension |
| 7490 | givinostat | To treat Duchenne muscular dystrophy in individuals aged 6 years and older |
n/i (not included_ as the agent does not meet the current criteria for inclusion in the GtoPdb.
Antibacterial Curation
Our collaboration with Antibiotic DB (ADB; www.antibioticdb.com) continues to allow us to extend the coverage of ligands with annotated antibacterial activity in GtoPdb and provide comprehensive chemistry and pharmacology for select antibacterials curated within ADB, via reciprocal links. This project is supported by the Global Antibiotic Research and Development Partnership (GARDP; https://gardp.org/).
Currently we have 537 ligands tagged in GtoPdb as ‘antibacterial’ and 512 of these have links to compounds at ADB. Since our last release we have added 30 new antibacterial ligands including:
- 6 drugs that are approved, or have been approved in the past, for clinical use in human
- 18 compounds that progressed to clinical evaluation
Website Updates
- Updated ligand substructure search to fix an issue with searches returning no results when explicit hydrogen atoms are included in the query structure.
- The search now opens a new tab for the results so that the original search tab with the query structure remains open and editable to support easier refinement by users of their query.
- Updates to our web services
- Antibacterial tag is now include as filter for ligands
- Interaction JSON extended to include target names, ligand names and selectivity
- Updated download files
- Updated data download files
- Approved drugs with their targets, including gene names, Entrez IDs and Ensembl IDs
- Interaction downloads split by target class
- Website
- Database content move from about page to it’s own page
















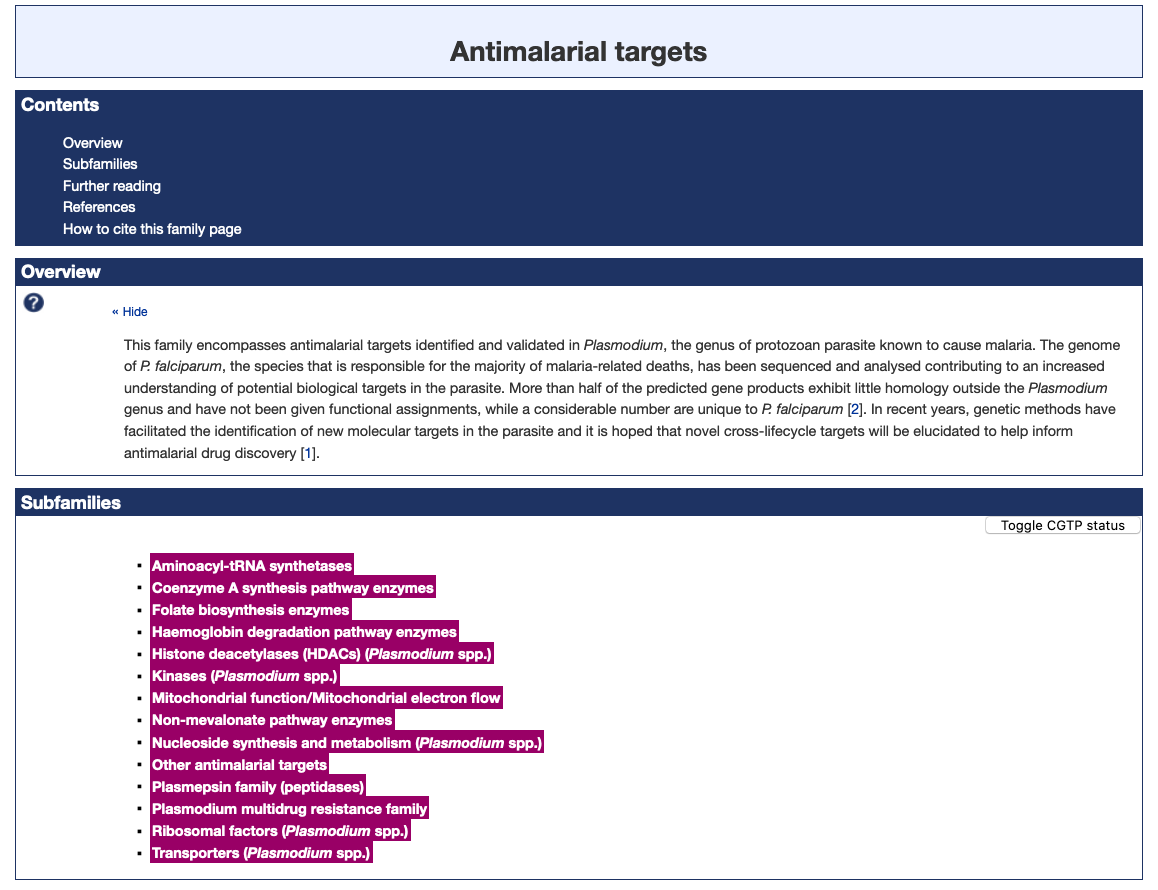 Figure 1. The Antimalarial targets family page illustrating the new subfamily classification (highlighted in magenta).
Figure 1. The Antimalarial targets family page illustrating the new subfamily classification (highlighted in magenta).








 In
In 




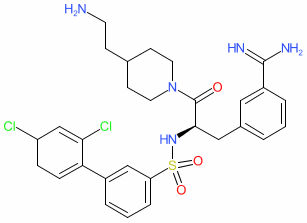
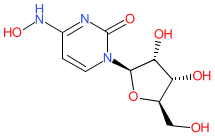





 We are delighted to announce the first full public release of the
We are delighted to announce the first full public release of the  The GtoMPdb portal homepage
The GtoMPdb portal homepage